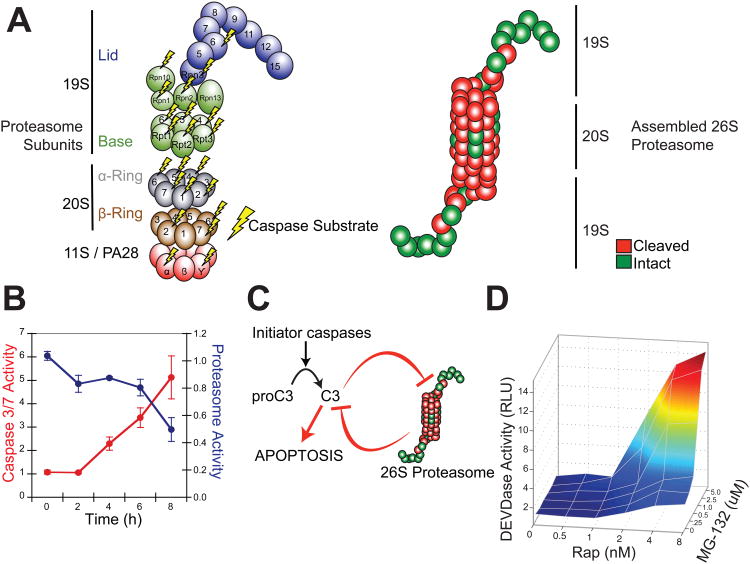
Figure 6. Reciprocal Negative Regulation between the 26S Proteasome and Executioner Caspases.
(A) Novel and published proteasome subunits identified as caspase substrates are mapped to an inventory of 20S core, 19S lid/base and 11S activator subunits (left). Cleaved subunits (red) are also mapped onto a model of the fully assembled, multimeric 26S proteasome (right). (B) The Casp3-TevS-2 line was treated with 10nM Rap over a course of 8 hr and assayed for caspase activity with Caspase-Glo-3/-7 and protease activity with Proteasome-Glo (Chymotrypsin-Like). Data are presented as the average from an experiment performed in triplicate (Error Bars±SD; note different scales). (C) A model for reciprocal negative regulation between activated executioner caspases and the 26S proteasome-ubiquitin system predicts synergy between caspase activation and proteasome inhibition. (D) The Casp3-TevS-2 line was co-treated with the indicated concentrations of Rap (0-8nM) and MG-132 (0-5uM) for 4 hr and caspase activation was measured with Caspase-Glo-3/-7 in triplicate. See also Table S1.