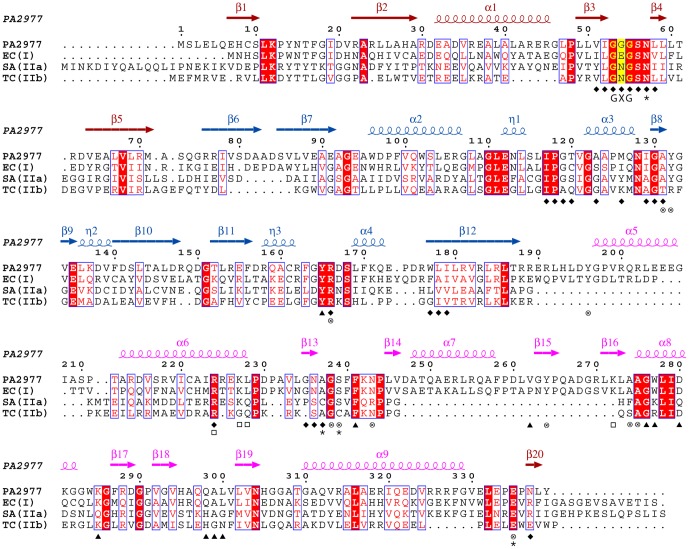
Figure 2. Structure-based sequence alignment of PaMurB against three MurB enzymes with experimentally determined crystal structures.
Each of the three enzymes represents one type of MurB. Annotations of secondary structure are based on PaMurB and are colored red for FAD-binding domain I, blue for domain II and pink for substrate-binding domain III. Strictly conserved residues are shaded in red and conserved regions are boxed; the GXG motif important for FAD-binding is indicated and shaded in yellow. The sequences compared are Pseudomonas aeruginosa MurB (PA2977), Escherichia coli MurB (EC(I), type I, PDB code 2MBR), Staphylococcus aureus MurB (SA(IIa), type IIa, PDB code 1HSK), and Thermus caldophilus MurB (TC(IIb), type IIb, PDB code 2GQT). Residues involved in cofactor- and substrate-binding are indicated: ⧫, residues that interact with FAD in both PaMurB and EcMurB; ▴, equivalent residues that interact with UNAGEP in EcMurB; □, residues that interact with NADP+;⊗, residues that interact with both UNAGEP and NADP+; ✶, residues that coordinate the putative catalytic metal ion.