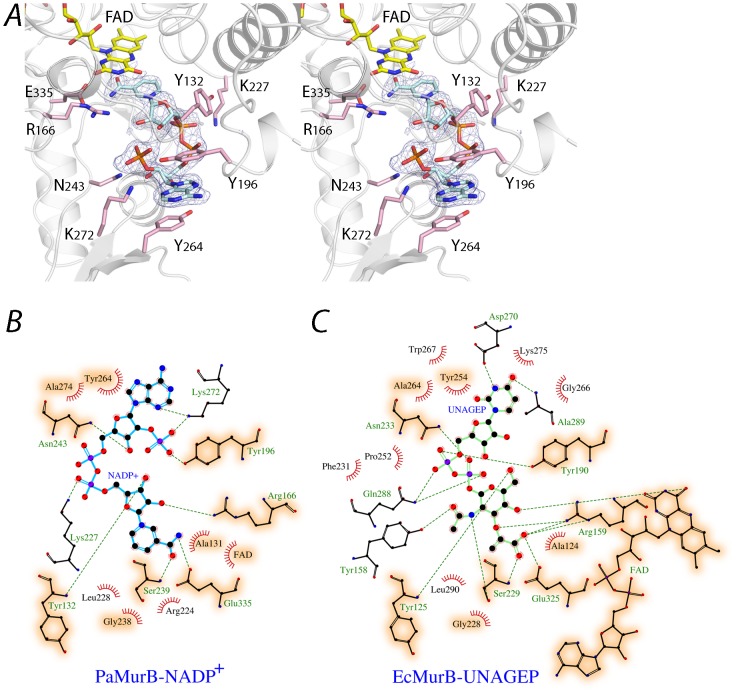
Figure 4. NADP+ and the substrate binding site.
A. Stereo view of NADP+ bound in the substrate channel of PaMurB. NADP+ is depicted as a cyan stick model. The Fo-Fc omit electron density of NADP+, contoured at 3.0 σ, indicates tight binding of the ligand. The nicotinamide ring stacks against the isoalloxazine ring system of FAD (shown in yellow). Residues of the binding site that form hydrogen bonds with NADP+ include Tyr-132, Arg-166 and Glu-335 for the nicotinamide moiety, Lys-227 for the diphosphate backbone, and Tyr-196, Asn-243 and Lys-272 for the adenosine. The adenosine moiety is in addition stabilized by stacking interactions with Tyr-196 and Tyr-264. B and C. Comparison of protein residues interacting with NADP+ and UNAGEP (based on EcMurB, PDB code 2MBR). NADP+ and UNAGEP are shown as ball-and-stick models in cyan and green, respectively. Red radiating lines around ligand atoms indicate van der Waals contacts, while green dashes represent potential hydrogen bonds. Ball-and-stick models are shown for binding site residues that provide polar interactions but not those involved in van der Waals interactions only. Orange shading highlights binding site residues that are conserved and involved in binding both substrates.