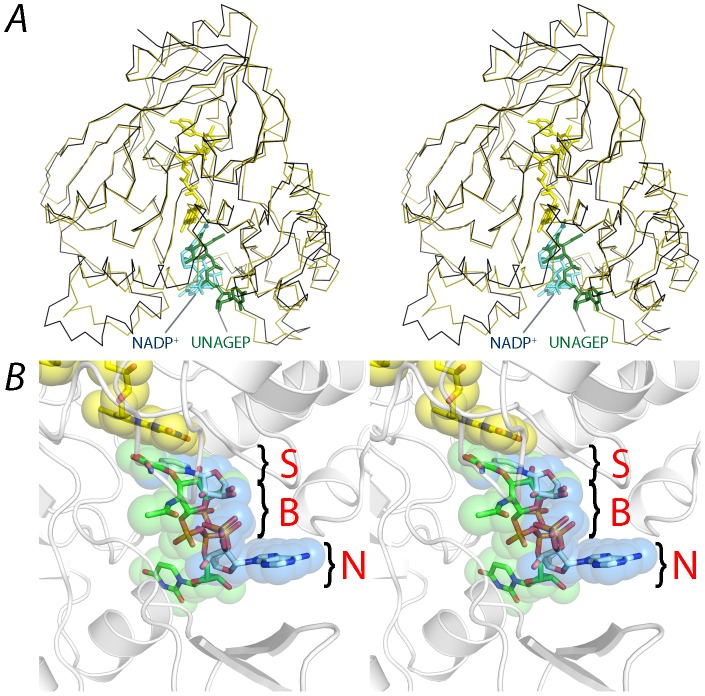
Figure 5. NADP+ shares the same substrate binding site with UNAGEP.
A. Stereo view of PaMurB (black ribbon) and UNAGEP-bound EcMurB (olive green ribbon, 2MBR) superimposed based on their FAD atomic coordinates. FAD, NADP+ and UNAGEP are depicted as stick models in yellow, cyan and green, respectively. NADP and UNAGEP occupy the same substrate channel located between the lobes of domain III. B. Co-localization of NADP(H) and UNAGEP substrate moieties on the si face of the FAD isoalloxazine ring system. The reactive moieties of the two substrates align together after superimposing the structures of PaMurB-NADP+ and EcMurB-UNAGEP (PDB code 2MBR) based on the FAD atomic coordinates. However, the non-reactive parts of the ligands diverge in the binding site, which can be visualized as three loci. S: the substrate moiety that reacts with FAD. B: the backbone region consisting of sugar and diphosphate. N: the non-reactive nucleotide moiety, which shows the greatest deviation. The remodeling of the binding site according to individual substrates is discussed in the text.