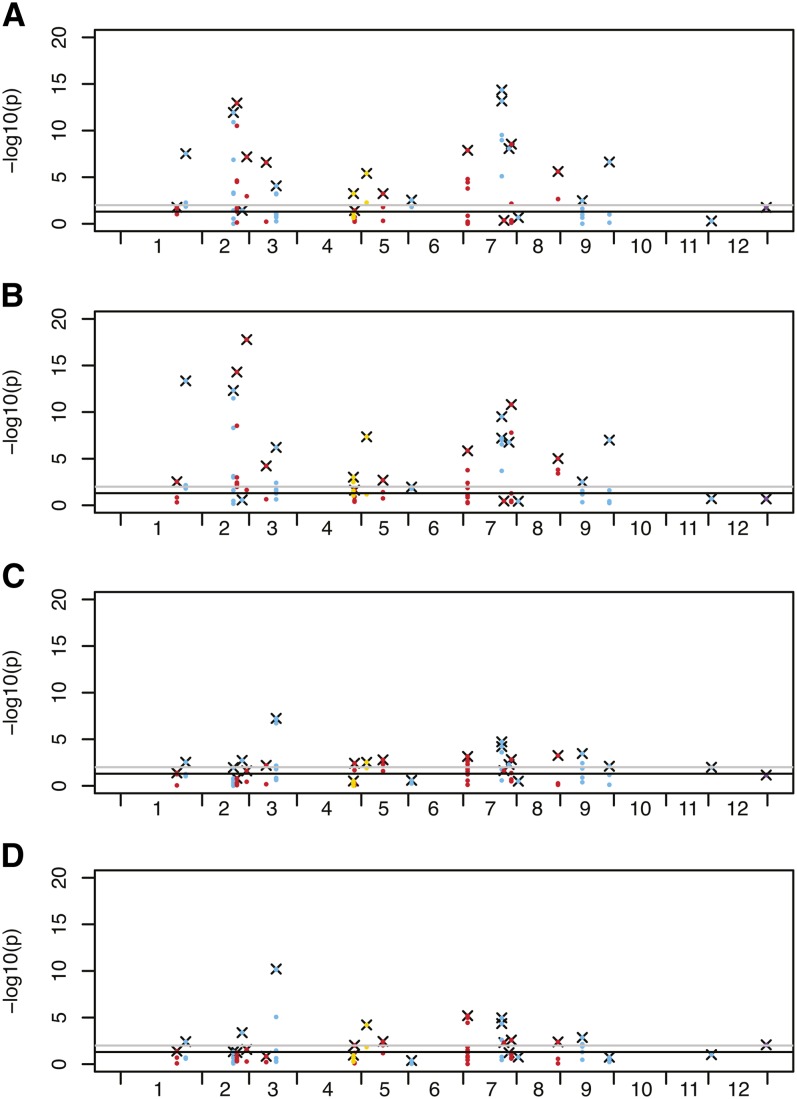
Figure 6.
Selection for carbohydrate synthesis, degradation, transport, and regulatory genes in Chip Processing clones. SNPs within 25 genes were tested for significant differences in allele and genotype composition between Chip Processing clones and all other cultivated potato [(A) allele, (B) genotype] and between French Fry Processing clones and non-processing cultivated potato clones [Pigmented, Round White Table, Table Russet, Yellow; (C) allele, (D) genotype]. The x-axis in all graphs is the position on each of the 12 potato chromosomes and the y-axis is a –log10 transformation of the P-value for the χ2 tests. Blue data points are SNPs in carbohydrate degradation genes, red data points are SNPs in carbohydrate synthesis genes, yellow data points are SNPs in carbohydrate transport genes, and the purple data point is a SNP in a regulatory gene annotated as invertase inhibitor. The data points with an x behind them are the most significant SNP within a gene. The black horizontal lines indicate significance at P = 0.05 and the gray lines at P = 0.01.