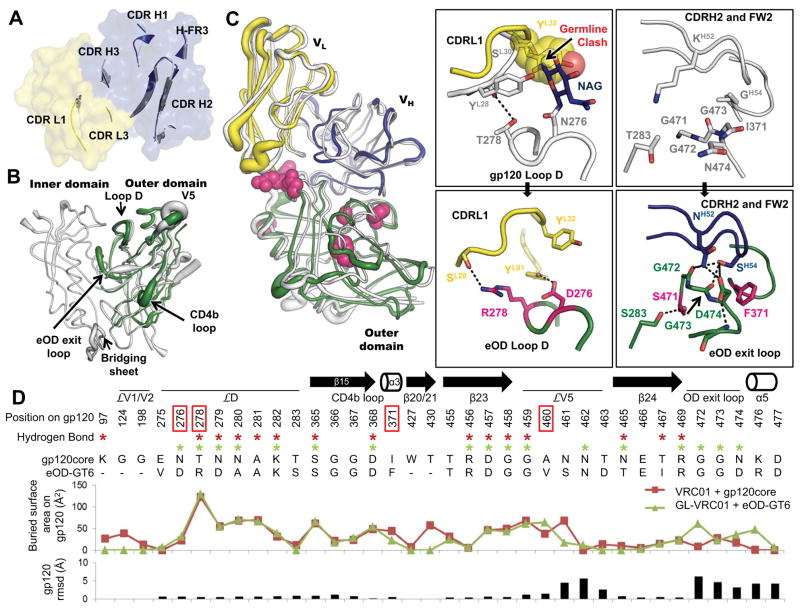
Fig. 2. Structural analysis of GL-VRC01 and eOD-GT6.
(A) Comparison between the crystal structure of an unliganded predicted GL-VRC01 antibody (heavy and light chains colored blue and yellow, respectively) and VRC01 bound to gp120 (gray). Variable regions of GL-VRC01 are rendered as a surface. Areas of GL-VRC01 and VRC01 that contact gp120, shown as secondary structure cartoons, are similar in conformation. (B) Comparison between the crystal structures of unliganded eOD-GT6 (green) and unliganded gp120 core from HIV-1 strain 93TH057 (PDBID: 3TGT, gray). Structures are rendered according to B-values, with thin and thick lines representing areas possessing low and high flexibility, respectively. (C) Comparison between the crystal structures of GL-VRC01+eOD-GT6 and VRC01+gp120 core. Components are colored as in (A) and (B) and shown as tubes. The positions of the recovered mutations in eOD-GT6 that enable binding of GL-VRC-class Abs are shown as space-fill magenta spheres. The angle of approach of GL-VRC01 and VRC01 to the CD4bs is nearly identical. Key areas where interactions are different between VRC01 on gp120 (upper panels) and GL-VRC01 on eOD-GT6 (bottom panels) are shown in insets. eOD-GT6 confers germline reactivity by removing a potential clash with the N276 glycan, as well as by creating additional contacts with loop D (inset I), the OD exit loop (inset II) and V5 (fig. S12). (D) gp120 residues involved in the VRC01+gp120 and GL-VRC01+eOD-GT6 interfaces are compared in sequence, H-bond (stars), surface buried area and RMSD. Interfaces were calculated using PDBePISA (29) and Cα rmsd using Chimera (30).