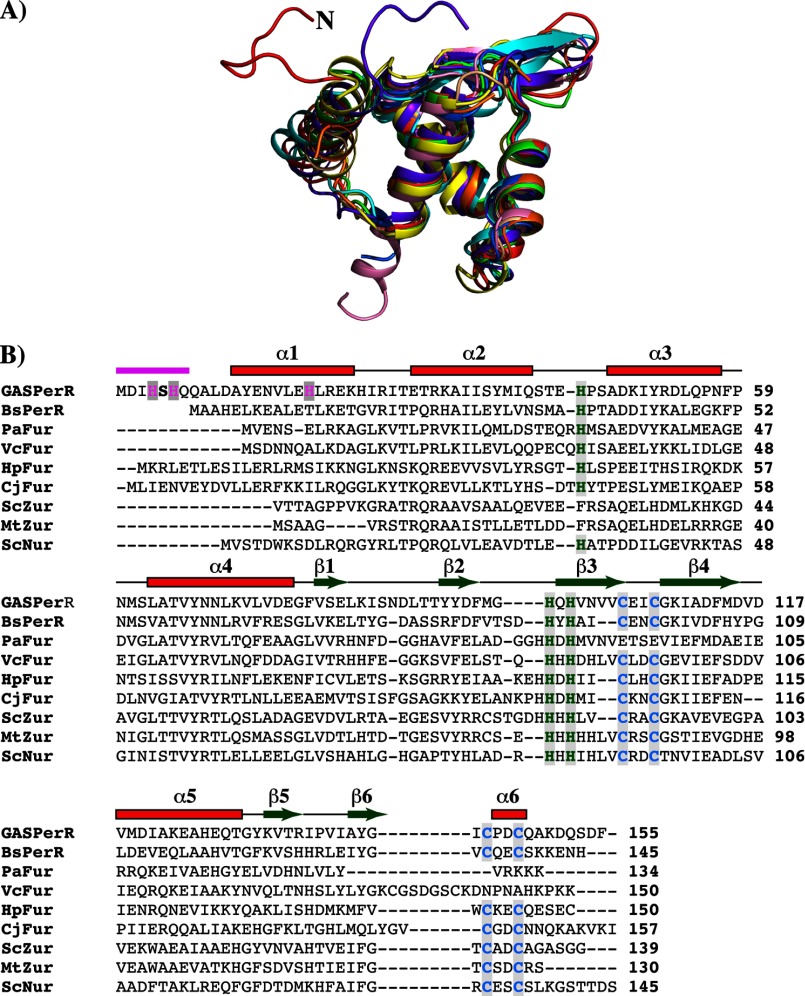
FIGURE 9.
N-terminal extension with the HXH metal-binding motif is unique in the structure of PerR. A, superposition of residues 2–94 of the N-terminal domain of PerR (red) with the analogous residues from the structures of PerRBs (green, PDB code 2FE3) (23) and other Fur regulators (Fur from Pseudomonas aeruginosa (orange, PDB code 1MZB), C. jejuni (yellow, PDB code 4ETS), H. pylori (purple, PDB code 2XIG), and Vibrio cholerae (light brown, PDB code 2W57); Zur from Streptomyces coelicolor (blue, PDB code 3MWM) and Mycobacterium tuberculosis (pink, PDB code 2O03); and Nur from Streptomyces coelicolor (cyan, PDB code 3EYY)) (43). The N-terminal extension of PerR is colored red and labeled (N). Structural superposition was performed with “LSQKAB”(47). B, amino acid sequence alignment of PerR with other structurally characterized Fur family regulators (B. subtilis PerR-BsPerR, Fur from P. aeruginosa, V. cholerae, H. pylori, and C. jejuni, PaFur, VcFur, HpFur, and CjFur, respectively; Zur from M. tuberculosis and S. coelicolor, MtZur and ScZur, respectively; and S. coelicolor Nur-ScNur). The multiple sequence alignment was performed with ClustalW (49). The secondary structure elements are labeled and marked above the alignment; α-helices are shown as rectangles and β-strands as arrows. The region corresponding to the N-terminal extension is shown in magenta. The highly conserved CXXC structural zinc motif is colored in blue, and the C-terminal metal-binding ligands are colored in green. The N-terminal HXH motif unique for PerR is highlighted in magenta.