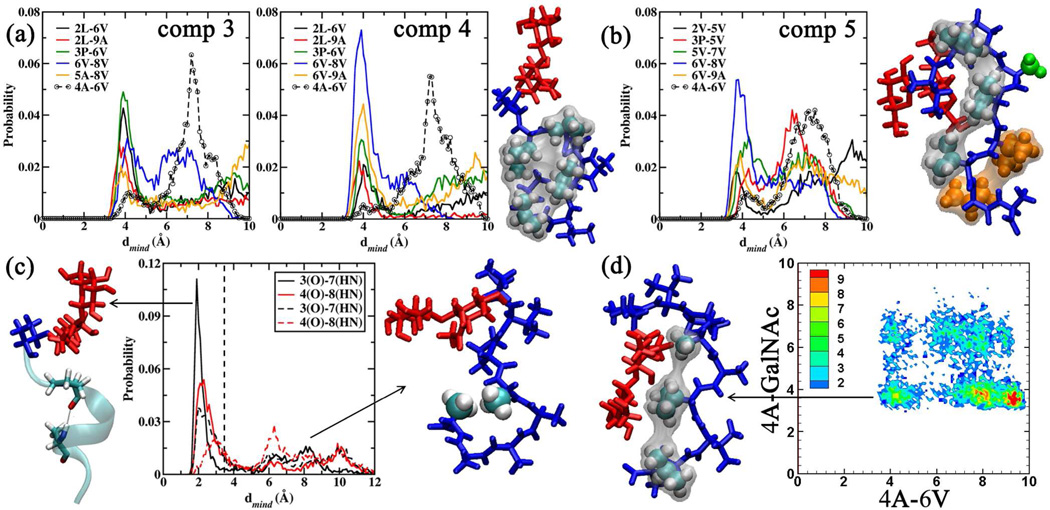
Figure 7.
(a) Close contact probabilities observed between the amino acids in 3 and 4. Representative structure for 4 obtained by clustering the conformations belonging to region I. Amino acids 2L, 3P, 6V and 8V are shown in VWD representation to highlight the hydrophobic clustering. (b) Close contact probabilities in 5. Representative structure for 5 obtained upon clustering conformations in region III. Amino acids 2V, 5V and 7V are presented in VWD representation with the corresponding surface to highlight the hydrophobic clustering. The other amino acids 4A (green VDW representation), 6V and 8V (orange VDW representation) highlight other features. (c) H-bond distribution for 3P(O)-7A(HN) (black lines) and 4A(O)-8A(HN) (red lines) for 9 (solid lines) and 11 (dashed lines). Representative structures for 9 and 11 depicting the favored α-helical folded and extended geometries. The peptide tail region is presented in new cartoon representation to highlight the α-helical fold for 9. Amino acids 4A and 8A are presented in VDW representation to highlight the hydrophobic clustering for 11. (d) 4A-6V/4A-GalNAc 2D distribution for 14.The representative structure corresponding to the collapsed region is presented. Amino acids 4A, 6V and 8V are presented in VDW representation. General Color Coding: Carbohydrate region is presented in red and the peptide region in blue. All distances are presented in Å.