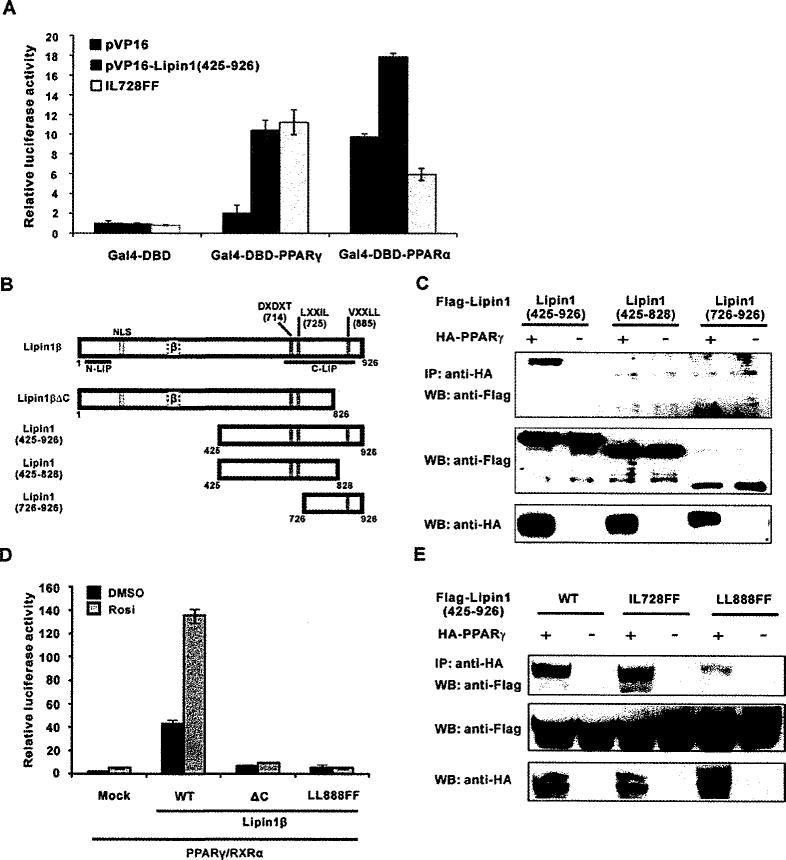
Figure 2. Lipin1 interacts with PPARγ via its C-terminal region.
(A and B) NIH 3T3 cells were transfected with the reporter plasmid pFR-Luc along with the vectors encoding GAL4-DBD–PPARα or –PPARγ. VP16–lipin1 (residues 425–927) or the IL728FF mutant construct were also co-transfected. Ligands for PPARα (WY14643) or PPARγ (rosiglitazone) was treated the next day and cells were incubated for another 24 h before the assay. (C) Lipin1 constructs used in the experiments. NLS, nuclear localization signal; DXDXT, PAP enzymatic motif; LXXIL, nuclear receptor-interaction motif; VXXLL, PPARγ-interacting motif. (D) NIH 3T3 cells were co-transfected with the 3× PPRE-tk luciferase construct and the PPARγ2 and RXR vectors, along with pSG5-KH2M1 (Mock), pSG5-lipin1β, pSG5-lipin1ΔC or pSG5-lipin1 LLBBBFF as indicated. The next day, cells were treated with 1 μM rosiglitazone (Rosi.) and incubated for another 24 h. Relative luciferase activity was calculated as described in Experimental section. (E) FLAG–triacylglycerolged lipin1 fragments or its mutants were overexpressed with or without HA–PPARγ in HE-K293 cells. Co-immunoprecipitation performed as described in Experimental section. IP, immunoprecipitation; WB, Western blot.