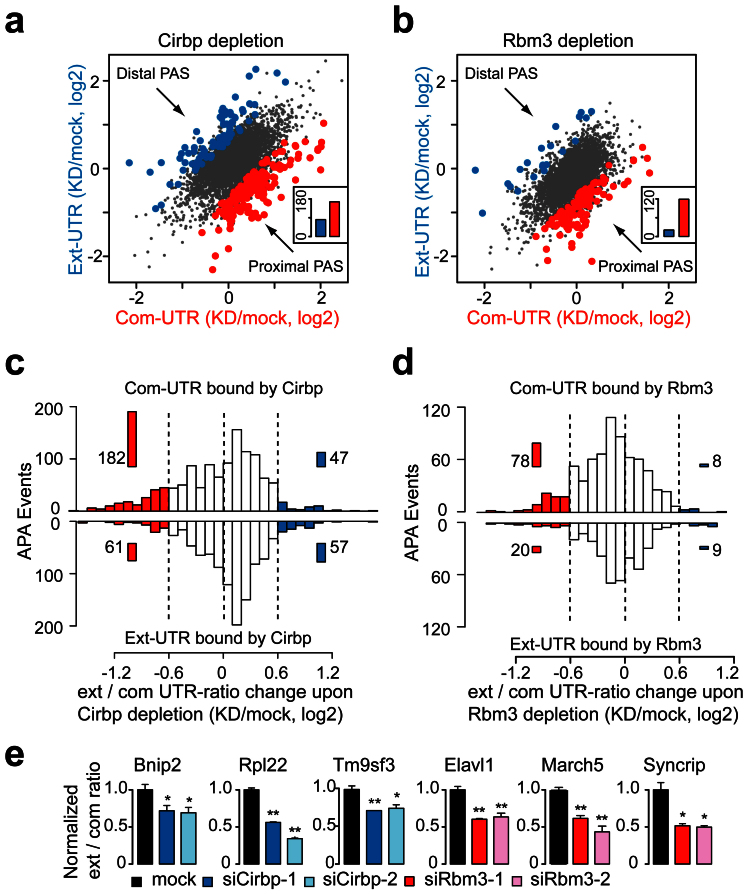
Figure 4. RNA-seq and PAR-CLIP analyses indicated that Cirbp and Rbm3 repressed the usage of proximal PASs by binding to the common 3′UTRs.
(a) Scatterplot of the common and extended 3′UTR expression of tandem UTR genes upon Cirbp depletion. Significantly increased expression of isoforms resulting from usage of distal (blue) and proximal PAS (red) are highlighted. Inset: total number of the different changes (Binomial test, P = 3.4E-08). (b) Scatterplot of the common and extended 3′UTRs expression of tandem UTR genes upon Rbm3 depletion as in (a) (Binomial test, P < 1.0E-15). (c) Histogram showed the ext/com UTR-ratio changes of the genes with Cirbp-binding sites in the tandem UTRs upon Cirbp depletion (Top, common 3′UTRs bound by Cirbp; Bottom, extended 3′UTRs bound by Cirbp). The PAS-usage switching events with up (blue) or downregulated (red) ext/com UTR-ratio were highlighted. Inset: the total number of cases in each region. (d) Histogram showed the ext/com UTR-ratio changes of the genes with Rbm3-binding sites in the tandem UTRs upon Rbm3 depletion as in (c). (e) QPCR analyses of the selected genes that showed downregulated ext/com UTR-ratio upon Cirbp or Rbm3 depletion. The statistical significance was calculated against the mock cells. See also Fig. S4G. Data are represented as mean +/- SEM (n = 3); Student's t-test * P < 0.05, ** P < 0.01.