Abstract
There is widespread agreement that cancer gene discovery requires high-quality tumor samples. However, whether primary tumors or cultured samples are superior for cancer genomics has been a longstanding subject of debate. This debate has recently become more important because federally funded cancer genomics has been centralized under The Cancer Genome Atlas, which has chosen to focus exclusively on primary tumors. Here, we provide a data-driven “perspective” on the effect of sample type selection on cancer genomics research. We show that, in the case of glioblastoma multiforme, primary tumors and xenografts are best for the identification of amplifications, whereas xenografts and cell lines are superior for the identification of homozygous deletions. We also note that many of the most important oncogenes and tumor suppressor genes have been discovered through the use of cell lines and xenografts, and highlight the lack of published evidence supporting the dogma that ex vivo culture generates artifactual genetic lesions. Based on this analysis, we suggest that cancer genomics projects such as The Cancer Genome Atlas should include a variety of sample types such as xenografts and cell lines in their integrated genomic analysis of cancer.
Introduction
After several decades in which cancer genomics research was performed in individual laboratories and funded by single-investigator grants, the field has recently been centralized and expanded under the auspices of The Cancer Genome Atlas (TCGA), which is performing integrated genomic analysis on a large number of samples from a wide range of common human tumor types. TCGA was initiated in December 2005, recently completed a 3-year pilot project [focused on glioblastoma multiforme (GBM), ovarian cancer, and lung cancer], and is currently organizing itself to begin the production phase of genomic analysis on a wider range of tumor types.
The procurement of high-quality cancer samples is the critical first step for cancer genomics projects such as TCGA. There are four principle types of human cancer samples available for such studies—primary tumors, primary cultures, primary xenografts, and established cell lines. The availability of each sample type is somewhat tumor type–specific (e.g., breast cancers do not efficiently form xenografts). Each of these sample types has unique advantages and disadvantages that are thought to affect the success of genomic analyses (see Supplementary Table S1).
Unlike other ongoing cancer genomics projects (1–3), TCGA has chosen to focus exclusively on the collection and analysis of primary tumor samples. This decision was based on considerations such as the fact that primary tumors can most easily be collected in large numbers in a prospective fashion, and the concern that ex vivo culture could induce artifactual genetic lesions. However, this decision was not based strictly on scientific data, as few (if any) published studies have directly evaluated the advantages and disadvantages of various sample types for genetic analysis.
We initially became interested in this issue of sample type selection for cancer genomics because, as TCGA was performing copy number analyses on GBM primary tumor samples (4), we were performing similar analyses on a panel of all four GBM sample types (5, 6). The results of these studies, described comprehensively for the first time in detail below, suggested that whereas primary tumors are an ideal sample type for the identification of genomic amplifications, they are inferior to xenografts and cell lines for the identification of genomic deletions. As such, this “perspective” will describe the effects of sample type on copy number analysis in GBM, examine the evidence supporting the widely accepted idea that cultured sample types contain artifactual genetic lesions, and review the role of different sample types in the history of cancer gene discovery.
Comparative Copy Number Analysis of Diverse GBM Sample Types
In an effort to experimentally address issues in sample type selection for cancer genomics projects, copy number analysis was performed on 58 GBM samples derived from all four GBM sample types—primary tumors, primary cultures, primary xenografts, and established cell lines.5 Copy number data from an additional panel of 50 cell lines were also analyzed.6
Initially, we identified amplifications and deletions of the major GBM oncogenes and tumor suppressor genes (Table 1A; Supplementary Table S2). There was a substantial discrepancy in the frequency of oncogene amplification between sample types. For example, amplification of EGFR was commonly found in primary tumors and xenografts, but rarely found in primary cultures and cell lines. This phenomenon of loss of amplifications in GBM cell lines has been previously described but was thought to be specific to EGFR (7, 8). However, our data indicate that amplification of other GBM oncogenes such as PDGFRA, CDK4, and MDM4 is similarly lost during in vitro culture, and suggest that primary tumors and xenografts are the best sample type for the identification of novel amplicons containing candidate oncogenes.
Table 1.
Significant sample type effects on copy number alterations in GBM
| (A) | ||||||
| 206 TCGA primary tumors |
12 primary tumors |
10 primary cultures |
15 xenografts | 21 cell lines |
50 Sanger CGP cell lines |
|
|
| ||||||
| High-level amplifications | ||||||
| EGFR | 43% | 83% | 10% | 40% | 5% | 2% |
| CDK4/6 | 16% | 8% | 0% | 13% | 5% | 4% |
| MDM2/4 | 15% | 17% | 10% | 13% | 10% | 4% |
| PDGFRA | 11% | 17% | 0% | 7% | 0% | 2% |
| CCND1/D2/D3 | 4% | 8% | 0% | 7% | 5% | 2% |
| Genomic deletions | ||||||
| CDKN2A/B | 55% | 58% | 60% | 87% | 81% | 70% |
| PTEN | 8% | 8% | 10% | 7% | 14% | 18% |
| CDKN2C | 3% | 0% | 10% | 27% | 19% | 20% |
| NF1 | 2% | 0% | 0% | 7% | 5% | 8% |
| (B) | ||||||
| Genomic deletions | ||||||
| CDKN2A | 1.0 ± 0.3 | 0.6 ± 0.1 | 0.3 ± 0.2 | 0.2 ± 0.1 | 0.3 ± 0.1 | |
| PTEN | 1.0 ± 0.3 | 0.7* | 0.2* | 0.2* | 0.5 ± 0.4 | |
| CDKN2C | 1.1 ± 0.3 | — | 0.4* | 1.1 ± 0.2 | 0.5 ± 0.3 | |
| NF1 | 1.4 ± 0.2 | — | — | 0.3* | 0.3* | |
NOTE: A, percentage of tumor samples with focal (<10 Mb) genomic deletion and high-level (copy number >7) focal amplification of the indicated gene loci. B, mean copy number and SD at the indicated gene loci in those tumor samples with focal genomic deletion. Two-tailed unpaired t test analysis was used to compare the statistical significance of any difference in frequency of copy number alteration (A) and mean copy number (B) between the TCGA primary tumors and other GBM tumor samples. Statistically significant differences (P < 0.05) in frequency (A) and copy number means (B) are highlighted in boldface. —, no samples with focal genomic deletion at the indicated gene loci.
, less than three samples with genomic deletion, no SD calculation or t test analysis possible.
Of note, this loss of oncogene amplification during tissue culture seems to be tumor type–specific, as there are examples of tumor types in which oncogenes are amplified at a similar frequency in both cultured and uncultured samples. For example, MYC or MYCN are amplified in 28 out of 37 neuroblastoma cell lines (76%),7 comparable to that observed in neuroblastoma primary tumor samples (9).
There was also a discrepancy in the frequency of identifiable tumor suppressor gene deletions between sample types. For example, deletions of the CDKN2A/B locus were identifiable in a much higher fraction of xenografts and cell lines than in primary tumors and primary cultures (Table 1A; Supplementary Table S2). Importantly, this disparity was not limited to CDK inhibitors, but was also present for PTEN, NF1, and PTPRD. In the case of PTPRD, deletions in primary tumors were very rarely identified, and therefore TCGA did not sequence the gene in their GBM pilot project (4). It was only the use of additional sample types that enabled the identification of frequent deletions and somatic mutations of this emerging tumor suppressor gene in GBM (6).
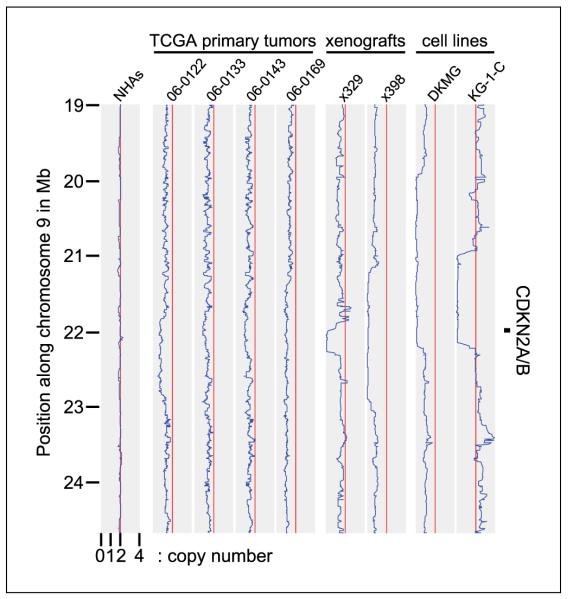
To determine whether the presence of admixed nonneoplastic cells and intratumoral genetic heterogeneity was responsible for impeding the identification of deletions in primary tumor samples, we analyzed CDKN2A/B and CDKN2C in both a first passage xenograft and the primary tumor from which it was derived. Deletions of both loci were present in the xenograft, but were largely masked in the primary tumor by the presence of admixed nonneoplastic cells and intratumoral genetic heterogeneity (5, 10). This same observation is evident when comparing copy numbers at each of the major tumor suppressor genes—deletions in primary tumors are more difficult to identify because their average copy number is significantly higher and their boundaries are less discrete (Table 1B; Fig. 1; Supplementary Table S3).
Figure 1.
Copy number plots along chromosome 9p for four TCGA primary tumors (reported to have homozygous deletion of CDKN2A/B), two xenografts, two cell lines, and normal human astrocytes (NHAs). Each of the depicted xenografts and cell lines have homozygous deletion of the CDKN2A/B locus with copy number <0.2, whereas the four TCGA primary tumors have hemizygous/heterogeneous deletion with copy numbers of 1.07, 1.32, 1.22, and 1.29 for TCGA-06-0122, TCGA-06-0133, TCGA-06-0143, and TCGA-06-0169 respectively.
Taken together, these data indicate that xenografts and cell lines are superior to primary tumors for the identification of genomic deletions. The presence of nonneoplastic cells and heterogeneity in even the most homogeneous tumor types such as GBM results in substantial “noise” in the analysis, which hinders the identification of deletions and leads to a high rate of false-negatives. Such noise would be expected to pose similar problems in other cancer genomics assays as well, including DNA sequencing.
No Evidence of Artifactual Genetic Lesions Caused by Ex vivo Culture
Many cancer researchers favor using primary tumors rather than cultured samples because of the widespread belief that ex vivo culture can lead to the accumulation of spurious genetic alterations. Concerns of this type reached a pinnacle 15 years ago, when there was substantial controversy about whether the recently identified deletions and mutations of the p16INK4a tumor suppressor gene could be artifacts of ex vivo culture (11, 12). After substantial high-profile debate, this concern was eventually refuted and it is now universally accepted that p16INK4a is one of the most commonly inactivated tumor suppressor genes in human cancer. However, such concerns remain firmly entrenched in the minds of most cancer researchers.
To test whether these concerns are valid, we catalogued all the copy number alterations present in each of our 58 samples. Strikingly, there were no examples of recurrent deletions or amplifications present exclusively in cultured samples. Additionally, if ex vivo culture specifically enriches for cells with deleted tumor suppressor genes, one would similarly expect culture to enrich for cells with amplified oncogenes. Yet as we show in Table 1A, ex vivo culture leads to a decrease in oncogene amplification in GBM cells, not the predicted increase.
Next, a comprehensive search of the literature was performed in an effort to identify studies that document copy number alterations and/or mutations present exclusively in cultured samples but not in primary tumors. Although we were able to identify several studies which showed expression differences between primary tumors and cultured samples (13, 14), we were unable to identify any studies documenting genetic lesions unique to cultured samples.
In contrast, Jones and colleagues recently provided remarkably strong evidence in support of the idea that cultured samples faithfully recapitulate the genetic profile present in the tumor from which they were derived. In their study, 287 of 289 mutations (99.3%) initially discovered in human colon cancer xenografts and cell cultures were similarly present in the primary tumors from which the cultured samples were derived (15). These data indicate that ex vivo culture of colon tumors does not lead to the formation or accumulation of spurious genetic aberrations.
Based on these findings, we believe that there is little convincing evidence to support the dogma that ex vivo culture leads to artifactual deletions, amplifications, and somatic mutations. As such, the risk of failing to identify deletions in human cancer samples due to an exclusive focus on primary tumors is likely to be substantially greater than the risk of identifying spurious genetic events by including other sample types in the analysis. This is especially true because it is relatively trivial to determine whether an event initially discovered in cultured samples is similarly present in primary tumors, as was the case, for example, with the recent identification of CDKN2C as a GBM tumor suppressor gene (5, 16).
Cultured Samples Have Been Used in the Discovery of Most Oncogenes and Tumor Suppressors
Finally, we looked back through the modern history of cancer genetics to identify the sample types used to discover the most commonly altered oncogenes and tumor suppressor genes (Table 2). Notably, most somatically altered cancer genes that were not discovered via linkage analysis were initially identified using xenografts and cell lines. This includes p53, PTEN, p16INK4a, K-Ras, PIK3CA, B-Raf, and others (11, 17–30). Based on this history, it seems prudent to include cultured samples in any cancer genomics initiative whose major goal is the identification of novel somatically altered cancer genes.
Table 2.
Sample types used in the initial discovery of major somatically altered cancer genes
| Gene | Tumor type(s) | Sample type(s) | Reference |
|---|---|---|---|
| Oncogenes | |||
| HRAS | Bladder carcinoma | Cell lines | (17) |
| KRAS | Colon carcinoma | Cell lines | (18) |
| NRAS | Neuroblastoma | Cell lines | (19) |
| MYC | Myeloid leukemia | Cell lines | (20) |
| EGFR | Glioma | Primary tumors | (21) |
| CTNNB1 (β-catenin) | Colon carcinoma | Cell lines | (22) |
| BRAF | Melanoma, others | Cell lines | (23) |
| PIK3CA | Colon carcinoma | Xenografts and primary cultures | (24) |
| Tumor suppressors | |||
| RB1 | Bladder carcinoma | Cell lines | (25, 26) |
| TP53 | Colon carcinoma | Xenografts | (27) |
| CDKN2A (p16INK4a) | Multiple | Cell lines | (11) |
| SMAD4 | Pancreatic carcinoma | Xenografts | (28) |
| PTEN | Multiple | Cell lines, xenografts, and primary cultures | (29, 30) |
Conclusions
Here, we provide three rationales for the inclusion of cultured samples in TCGA and other cancer genomics efforts. First, we show that in the case of one major human tumor type, there are significant differences in the utility of different sample types for the identification of copy number alterations. Second, we document that there is little evidence supporting the popular notion that ex vivo culture of human tumors leads to spurious genetic alterations. And third, we show that most major somatically altered cancer genes discovered to date were identified using xenografts and cell lines. Based on these arguments, we believe it would be prudent for TCGA to include a range of sample types in their burgeoning analysis of cancer genomics. We also note that the use of cultured samples is supported by the Cancer Genome Project of the Wellcome Trust Sanger Institute and is within the agreed guidelines of the International Cancer Genome Consortium.
Supplementary Material
Acknowledgments
Grant support: National Cancer Institute, American Cancer Society, and Georgetown University School of Medicine (T. Waldman).
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
These data were generated using Affymetrix 250K NspI SNP arrays and analyzed using dChip, a publicly available software program (http://biosun1.harvard.edu/complab/dchip/). These data have been reported on previously (5, 6), and the raw and processed data sets have been deposited into the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/), accession number GSE13021.
Copy number data for a panel of 50 malignant glioma cell lines using Affymetrix SNP 6.0 arrays was generated by the Cancer Genome Project of the Wellcome Trust Sanger Institute, and is publicly available at http://www.sanger.ac.uk/genetics/CGP.
Copy number data for a panel of 37 neuroblastoma cell lines using Affymetrix SNP 6.0 arrays was generated by the Cancer Genome Project of the Wellcome Trust Sanger Institute, and is publicly available at http://www.sanger.ac.uk/genetics/CGP.
Disclosure of Potential Conflicts of Interest No potential conflicts of interest were disclosed.
References
- 1.Greenman C, Stephens P, Smith R, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–8. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parsons DW, Jones S, Zhang X, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–12. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jones S, Zhang X, Parsons DW, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–6. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.The Cancer Genome Atlas Research Network Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–8. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Solomon DA, Kim JS, Jenkins S, et al. Identification of p18INK4c as a tumor suppressor gene in glioblastoma multiforme. Cancer Res. 2008;68:2564–9. doi: 10.1158/0008-5472.CAN-07-6388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Solomon DA, Kim JS, Cronin JC, et al. Mutational inactivation of PTPRD in glioblastoma multiforme and malignant melanoma. Cancer Res. 2008;68:10300–6. doi: 10.1158/0008-5472.CAN-08-3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bigner SH, Humphrey PA, Wong AJ, et al. Characterization of the epidermal growth factor receptor in human glioma cell lines and xenografts. Cancer Res. 1990;50:8017–22. [PubMed] [Google Scholar]
- 8.Pandita A, Aldape KD, Zadeh G, Guha A, James CD. Contrasting in vivo and in vitro fates of glioblastoma cell subpopulations with amplified EGFR. Genes Chromosomes Cancer. 2004;39:29–36. doi: 10.1002/gcc.10300. [DOI] [PubMed] [Google Scholar]
- 9.Brodeur GM, Seeger RC, Schwab M, Varmus HE, Bishop JM. Amplification of N-myc in untreated human neuroblastomas correlates with advanced disease stage. Science. 1984;224:1121–4. doi: 10.1126/science.6719137. [DOI] [PubMed] [Google Scholar]
- 10.Solomon DA, Kim JS, Jean W, Waldman T. Conspirators in a capital crime: co-deletion of p18INK4c and p16INK4a/p14ARF/p15INK4b in glioblastoma multiforme. Cancer Res. 2008;68:8657–60. doi: 10.1158/0008-5472.CAN-08-2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kamb A, Gruis NA, Weaver-Feldhaus J, et al. A cell cycle regulator potentially involved in genesis of many tumor types. Science. 1994;264:436–40. doi: 10.1126/science.8153634. [DOI] [PubMed] [Google Scholar]
- 12.Cairns P, Mao L, Merlo A, et al. Rates of p16 (MTS1) mutations in primary tumors with 9p loss. Science. 1994;265:415–6. doi: 10.1126/science.8023167. [DOI] [PubMed] [Google Scholar]
- 13.Camphausen K, Purow B, Sproull M, et al. Influence of in vivo growth on human glioma cell line gene expression: convergent profiles under orthotopic conditions. Proc Natl Acad Sci U S A. 2005;102:8287–92. doi: 10.1073/pnas.0502887102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stein WD, Litman T, Fojo T, Bates SE. A serial analysis of gene expression (SAGE) database analysis of chemosensitivity: comparing solid tumors with cell lines and comparing solid tumors from different origins. Cancer Res. 2004;64:2805–16. doi: 10.1158/0008-5472.can-03-3383. [DOI] [PubMed] [Google Scholar]
- 15.Jones S, Chen WD, Parmigiani G, et al. Comparative lesion sequencing provides insights into tumor evolution. Proc Natl Acad Sci U S A. 2008;105:4283–8. doi: 10.1073/pnas.0712345105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wiedemeyer R, Brennan C, Heffernan TP, et al. Feedback circuit among INK4 tumor suppressors constrains human glioblastoma development. Cancer Cell. 2008;13:355–64. doi: 10.1016/j.ccr.2008.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taparowsky E, Suard Y, Fasano O, Shimizu K, Goldfarb M, Wigler M. Activation of the T24 bladder carcinoma transforming gene is linked to a single amino acid change. Nature. 1982;300:762–5. doi: 10.1038/300762a0. [DOI] [PubMed] [Google Scholar]
- 18.McCoy MS, Toole JJ, Cunningham JM, Chang EH, Lowy DR, Weinberg RA. Characterization of a human colon/lung carcinoma oncogene. Nature. 1983;302:79–81. doi: 10.1038/302079a0. [DOI] [PubMed] [Google Scholar]
- 19.Shimizu K, Goldfarb M, Suard Y, et al. Three human transforming genes are related to the viral ras oncogenes. Proc Natl Acad Sci U S A. 1983;80:2112–6. doi: 10.1073/pnas.80.8.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Collins S, Groudine M. Amplification of endogenous myc-related DNA sequences in a human myeloid leukaemia cell line. Nature. 1982;298:679–81. doi: 10.1038/298679a0. [DOI] [PubMed] [Google Scholar]
- 21.Libermann TA, Nusbaum HR, Razon N, et al. Amplification, enhanced expression and possible rearrangement of EGF receptor gene in primary human brain tumors of glial origin. Nature. 1985;313:144–7. doi: 10.1038/313144a0. [DOI] [PubMed] [Google Scholar]
- 22.Morin PJ, Sparks AB, Korinek V, et al. Activation of β-catenin-Tcf signaling in colon cancer by mutations in β-catenin or APC. Science. 1997;275:1787–90. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- 23.Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–54. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 24.Samuels Y, Wang Z, Bardelli A, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 25.Friend SH, Bernards R, Rogelj S, et al. A human DNA segment with properties of the gene that predisposes to retinoblastoma and osteosarcoma. Nature. 1986;323:643–6. doi: 10.1038/323643a0. [DOI] [PubMed] [Google Scholar]
- 26.Horowitz JM, Yandell DW, Park SH, et al. Point mutational inactivation of the retinoblastoma antioncogene. Science. 1989;243:937–40. doi: 10.1126/science.2521957. [DOI] [PubMed] [Google Scholar]
- 27.Baker SJ, Fearon ER, Nigro JM, et al. Chromosome 17 deletions and p53 gene mutations in colorectal carcinomas. Science. 1989;244:217–21. doi: 10.1126/science.2649981. [DOI] [PubMed] [Google Scholar]
- 28.Hahn SA, Schutte M, Hoque AT, et al. DPC4, a candidate tumor suppressor gene at human chromosome 18q21.1. Science. 1996;271:350–3. doi: 10.1126/science.271.5247.350. [DOI] [PubMed] [Google Scholar]
- 29.Li J, Yen C, Liaw D, et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997;275:1943–7. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- 30.Steck PA, Pershouse MA, Jasser SA, et al. Identification of a candidate tumor suppressor gene, MMAC1, at chromosome 10q23.3 that is mutated in multiple advanced cancers. Nat Genet. 1997;15:356–62. doi: 10.1038/ng0497-356. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.