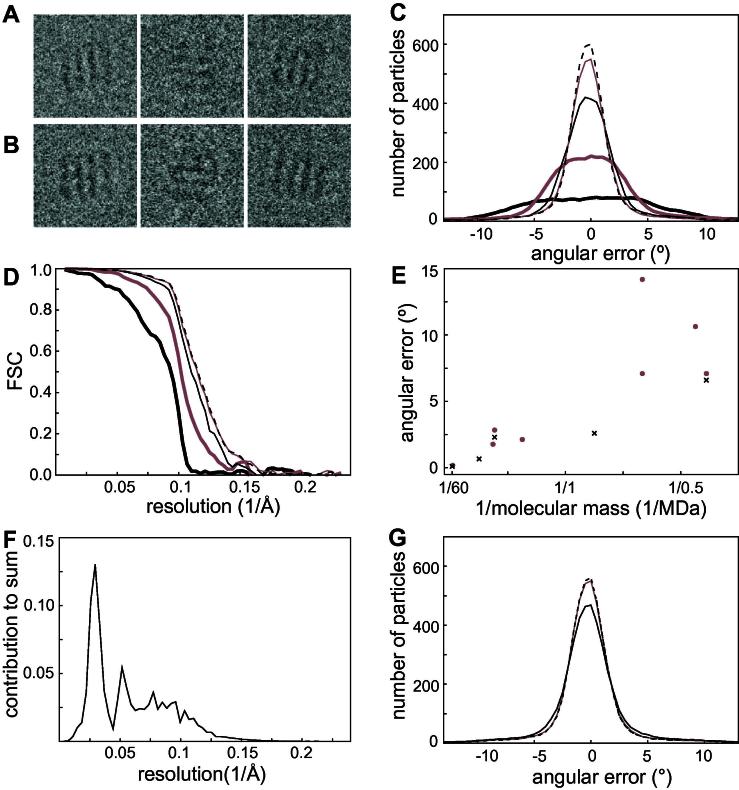
Fig.2.
Assessment of angular accuracies. (A) Three simulated GroEL particles. (B) The experimental counterparts of the particles in A. (C) Distribution of the angular errors after a single iteration of refinement of a 10 Å low-pass filtered version of the phantom against the simulated data, using an angular sampling rate of (bold black), (bold grey), (solid black), (solid grey) or (dashed black). (D) FSC with the phantom for the reconstructions from the refinements in C. (E) Experimentally determined angular accuracies based on tilt pair analysis (grey circles) compared to estimated angular accuracies based on the criterion (black crosses). The samples analyzed by tilt-pair analysis were rotavirus double-layered particle (50 MDa), chicken anemia virus (2.7 MDa), 70S ribosome (2.7 MDa), fatty acid synthase (2.6 MDa), pyruvate dehydrogenase (1.6 MDa), V and F-type ATPase (0.6 MDa), DNA-dependent protein kinase (0.47 MDa) and β-galactosidase (0.45 MDa). The specimens analyzed in RELION were rotavirus recoated particle (60 MDa), hepatitis B capsid (4 MDa), 70S ribosome (2.7 MDa), GroEL (0.8 MDa) and β-galactosidase (0.45 MDa). (F) Contribution of the different resolution shells to the summation inside the exponential in Eq. (8) for projections of the GroEL phantom with an angular distance of 2.7° between and . (G) Angular error distributions after alignment of the simulated GroEL particles against the phantom map using an angular sampling of 1.8°. The maximum resolution used in the alignment was varied between 20 Å (solid black), 10 Å (solid grey) and Nyquist (dashed black).