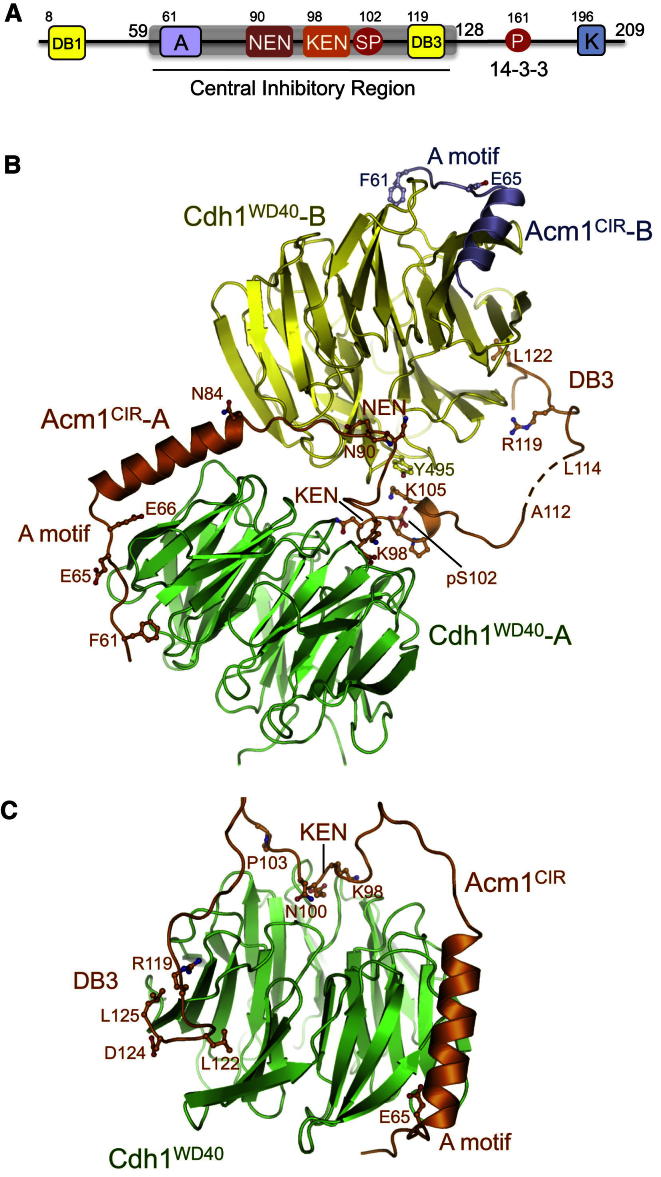
Figure 1.
Structure of Acm1CIR-Cdh1WD40
(A) Schematic of S. cerevisiae Acm1. A, A motif; DB1, N-terminal D box; NEN, 90N-91E-92N; KEN and DB3, pseudosubstrate KEN and D box (DB3) inhibitory motifs; SP, S102 CDK2 phosphorylation site; P, pT161-14-3-3 binding site; K, K motif.
(B) Acm1CIR-Cdh1WD40 heterotetramer. F113 of Acm1CIR-A was not located in the electron density maps and is represented with a dashed line.
(C) Model for the Acm1CIR-Cdh1WD40 heterodimer. A 13 residue linker was modeled between P103 (KEN) and R119 (DB3) (separated by 15 residues), indicating that it is stereochemically possible for the KEN box and DB3 motif of Acm1 to interact with their respective sites on the same Cdh1 molecule. K123 of the DB3 was omitted for clarity. See also Figure S1.