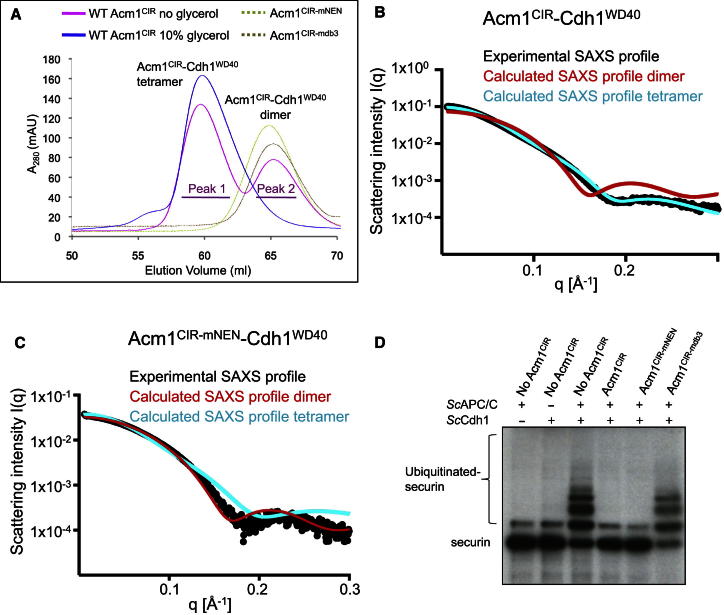
Figure 6.
Oligomeric States of Acm1-Cdh1WD40
(A) Size-exclusion chromatography of WT Acm1CIR-Cdh1WD40 without glycerol shows heterotetramers (peak 1) and heterodimers (peak 2). Glycerol (10% v/v) promotes heterotetramers. Associated SDS-PAGE gels are shown in Figures S4B and S4D. Only heterodimers are observed when either the NEN sequence (Acm1CIR-mNEN-Cdh1WD40) or DB3 is disrupted (Acm1CIR-mdb3-Cdh1WD40). The latter two samples were run with 10% (v/v) glycerol. Associated SDS-PAGE gels shown in Figures S4I and S4J.
(B and C) SAXS profiles for Acm1CIR-Cdh1WD40 and Acm1CIR-mNEN-Cdh1WD40 complexes. (B) Experimental SAXS profile (black dots) for Acm1CIR-Cdh1WD40 compared with computed SAXS profiles for the Acm1CIR-Cdh1WD40 tetramer (cyan) and dimer (red). It shows a good fit to the computed SAXS profile of the tetramer (χ2 = 12) but is a poor match to the computed SAXS profile of the dimer (χ2 = 93). The experimental radius of gyration (Rg) for Acm1CIR-Cdh1WD40 is 31 Å, and the calculated radii of gyration for tetramer and dimer models are 28 Å and 20 Å, respectively. The scattering vector q = 4πsin(θ)/λ, where θ is half the scattering angle. (C) Experimental SAXS profile (black dots) of Acm1CIR-mNEN-Cdh1WD40 shows a good fit to the computed SAXS profile of the dimer (χ2 = 3.7) but is a poor match to the computed SAXS profile of the tetramer (χ2 = 9.6). Experimental radius of gyration is 25 Å.
(D) In vitro APC ubiquitination assays with 35S-labeled S. cerevisiae securin and IVT-produced ScCdh1 and WT and mutant Acm1CIR. Acm1CIR-mediated inhibition is abolished by mutation of the DB3 (Acm1CIR-mdb3). In contrast to DB3 mutations, disruption of the NEN sequence (Acm1CIR-mNEN) does not impair Acm1CIR-mediated inhibition of APC/CCdh1 ubiquitination of securin.