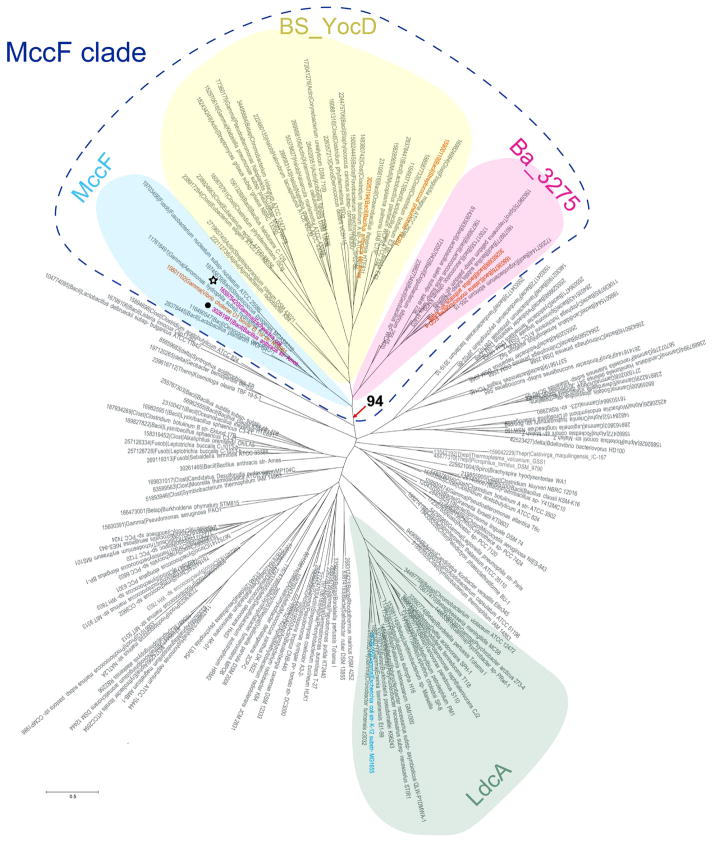
Figure 7.
Phylogenetic tree of S66 family of serine peptidases. The tree was constructed using confidently aligned blocks corresponding to conserved regions of multiple alignments of the S66 family of serine peptidases (174 sequences in total, 188 aligned positions; see Supporting material Fig. S1 for details). Three branches of the MccF clade and LcdA branch are shaded. The bootstrap value for the MccF clade is indicated. The EcMccF(black star) and BaMccF (black dot) sequences are indicated by magenta; sequences corresponding to proteins with determined substrate specificity are indicated by orange. Each terminal node of the tree is labeled by the Genbank Identifier (GI) number, five-letter taxonomy code of an organism and full systematic name of an organism. The taxonomy code is as follows: Gamma – Gammaproteobacteria; Beta – Betaproteobacteria; Alpha - Alphaproteobacteria; delta - Deltaproteobacteria; Bacil – Bacilli; Clost – Clostridia; Deino - Deinococcus-Thermus group; Syner - Synergistetes; Therm - Thermotogae; Acido - Acidobacteria group; Gemma - Gemmatimonadetes; Chroo – Chroococcales; Oscil – Oscillatoriales; Proch - Prochlorales; Nosto -Nostocales; Molli – Mollicutes; Spiro - Spirochaetes; Halob – Halobacteria; Actin – Actinobacteria; Fusob – Fusobacteria; Fibro – Fibrobacteres; Bacte - Bacteroidetes; Negat - Negativicutes; Thepl – Thermoplasmatales; Thepr – Thermoproteales.