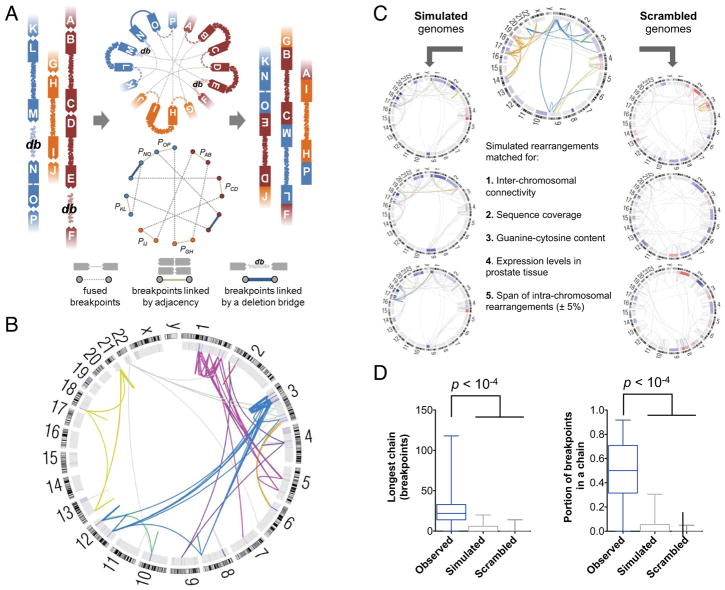
Figure 3. The ChainFinder Algorithm.
(A) ChainFinder creates a graph representation of genomic breakpoints that may be linked in chains by somatic fusions, statistical adjacency or deletion bridges. ChainFinder assigns two neighboring breakpoints to the same chain if the p-value for their independent generation (P) is rejected with a false-discovery rate below 10−2. For each cycle (closed path) within the graph, all scenarios are considered where one or more rearrangements in the cycle could have arisen independently. All rearrangements in a cycle are assigned to the same chain if every such scenario is rejected with a family-wise error rate below 10−2 across all scenarios. Please see the Supplemental Experimental Procedures for additional details.
(B) Circos plot of chained rearrangements in a prostate adenocarcima (P09-1042). Rearrangements depicted in the same color arose within the same chain; fusions in gray were not assigned to a chain. The inner ring depicts copy number gains and losses in blue and red, respectively.
(C) The false positive rate of ChainFinder was assessed using simulated and scrambled genomes based on observed rearrangements.
(D) For observed, simulated and scrambled genomes, the longest chain was compared along with the portion of breakpoints in any chain. Median values, middle quartiles, and range are indicated.
See also Figure S3.