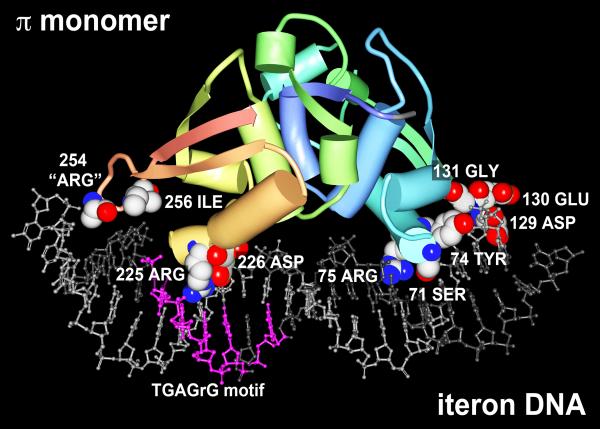
Figure 2.
A model for the interaction of a π protein monomer with R6K γ ori iteron DNA. The crystal structure for a variant π monomer bound to an iteron was solved by Swan et al. (2006) and deposited as a PDB file (PDB ID: 2NRA) to the Worldwide Protein Data Bank (www.pdb.org). The model shown was rendered using Protein Workshop 3.9 software (Moreland et al., 2005) and colors were enhanced using Adobe Photoshop 11.0.2. DNA strands (ball and stick representation) are light & dark gray with the TGAG(n)G motif in fuchsia. Amino acids predicted and/or shown to contact bases of the DNA (Swan et al., 2006; Kunnimalaiyaan et al., 2007, Saxena et al., 2010b) and shown in CPK are: Ser71, Tyr,74, Arg75, Asp129, Glu130, Gly131, Arg225, Asp226, “Arg254”, and Ile256. The PDB file does not account for Arg254 in its entirety. Atoms of CPK amino acids are colored as follows: C is white, N is blue, O is red. The remainder of the π monomer is represented as a chain color ramp from blue (start) to red (end) with helices and strands represented as cylinders and arrows, respectively.