Abstract
AIM: To investigate the expression patterns of long non-coding RNAs (lncRNAs) in gastric cancer.
METHODS: Two publicly available human exon arrays for gastric cancer and data for the corresponding normal tissue were downloaded from the Gene Expression Omnibus (GEO). We re-annotated the probes of the human exon arrays and retained the probes uniquely mapping to lncRNAs at the gene level. LncRNA expression profiles were generated by using robust multi-array average method in affymetrix power tools. The normalized data were then analyzed with a Bioconductor package linear models for microarray data and genes with adjusted P-values below 0.01 were considered differentially expressed. An independent data set was used to validate the results.
RESULTS: With the computational pipeline established to re-annotate over 6.5 million probes of the Affymetrix Human Exon 1.0 ST array, we identified 136053 probes uniquely mapping to lncRNAs at the gene level. These probes correspond to 9294 lncRNAs, covering nearly 76% of the GENCODE lncRNA data set. By analyzing GSE27342 consisting of 80 paired gastric cancer and normal adjacent tissue samples, we identified 88 lncRNAs that were differentially expressed in gastric cancer, some of which have been reported to play a role in cancer, such as LINC00152, taurine upregulated 1, urothelial cancer associated 1, Pvt1 oncogene, small nucleolar RNA host gene 1 and LINC00261. In the validation data set GSE33335, 59% of these differentially expressed lncRNAs showed significant expression changes (adjusted P-value < 0.01) with the same direction.
CONCLUSION: We identified a set of lncRNAs differentially expressed in gastric cancer, providing useful information for discovery of new biomarkers and therapeutic targets in gastric cancer.
Keywords: Long non-coding RNA, Gastric cancer, Microarray analysis, Data mining
Core tip: Long non-coding RNAs (lncRNAs) have risen to prominence with important roles in a broad range of biological processes. LncRNA expression patterns and their biological functions in gastric cancer still remain unknown. We re-annotated the probes from an Affymetrix Human Exon 1.0 ST array and identified probes uniquely mapping to lncRNAs at the gene level. These probes correspond to 9294 lncRNAs, covering nearly 76% of the GENCODE lncRNA data set. We identified a set of lncRNAs that were differentially expressed in gastric cancer. In an independent data set, 59% of these differentially expressed lncRNAs showed significant expression changes with the same direction.
INTRODUCTION
Over the last decade, advances in genome-wide analysis of gene expression have revealed far more genomic transcription than previously anticipated, with the majority of the genome being transcribed into non-coding RNAs (ncRNAs)[1,2]. Much attention has focused on microRNAs (miRNAs), one class of small non-coding RNAs. MiRNAs are involved in specific regulation of both protein-coding and putatively non-coding genes by post-transcriptional silencing or infrequently by activation[3-5].
More recently, long non-coding RNAs (lncRNAs), generally defined as having a size greater than 200 nucleotides, have risen to prominence with important roles in a broad range of biological processes. LncRNAs regulate gene expression at the level of post-transcriptional processing such as protein synthesis, RNA maturation, and transport. They also exert their effects in transcriptional gene silencing through the regulation of chromatin structure[6,7]. Dysregulation of lncRNAs is associated with many human diseases, including various types of cancers[8]. The well-studied lncRNA HOTAIR, for example, was found to be highly upregulated in both primary and metastatic breast tumors, and its expression level in primary tumors was a powerful predictor of eventual metastasis and death[9]. However, lncRNA expression patterns and their biological function in gastric cancer remain unknown.
In this study, we identified a set of lncRNAs that were differentially expressed in gastric cancer by analyzing publicly available data sets from the gene expression omnibus (GEO).
MATERIALS AND METHODS
Microarray data
Human exon arrays for gastric cancer and normal adjacent tissue were downloaded from the GEO. Two data sets were included: GSE27342 and GSE33335. GSE27342 consisted of 80 paired gastric cancer and normal adjacent tissue, including 4 stage I, 7 stage II, 54 stage III and 7 stage IV[10,11]. All samples were taken from three hospitals affiliated with Jilin University College of Medicine and Jilin Provincial Cancer Hospital, Changchun, China. GSE33335 consisted of 25 paired gastric cancer and normal adjacent tissue obtained from the tissue bank of Shanghai Biochip Center, Shanghai, China[12,13]. Three raw CEL files failed to be normalized and were excluded from our analysis, leaving 22 paired gastric cancer and normal adjacent tissue. GSE27342 was used as an experimental set to discover differentially expressed lncRNAs in gastric cancer while GSE33335 was used as a validation set.
Probe re-annotation pipeline
The sequences of protein-coding transcripts were retrieved from Ensembl release 67[14], UCSC[15] and RefSeq release 54[16] in July 2012. Specifically, the protein-coding transcripts are a pool of transcripts with gene_type as “protein_coding” in Ensembl, transcripts with category as “coding” in UCSC and transcripts with an identifier beginning with NM_ in RefSeq. The sequences of non-coding transcripts were compiled from Ensembl through Biomart. The probe sequences of the human exon array were downloaded from the Affymetrix website (http://www.affymetrix.com/Auth/analysis/downloads/na25/wtexon/HuEx-1_0-st-v2.probe.tab.zip) and aligned to the sequences of protein-coding and non-coding transcripts using BLAST-2.2.26+[17]. The alignment results were then filtered by the following steps: (1) probes perfectly matched to a transcript were retained; (2) probes mapped to non-coding transcripts only were retained; (3) probes mapped to unique genes were retained; (4) probes mapped to known lncRNAs (genes annotated with processed_transcript, lincRNA, antisense, non_coding, sense_intronic, ncrna_host, sense_overlapping and 3pri-me_overlapping_ncrna) were retained; and (5) genes with less than 3 probes were removed.
A new PGF file covering the re-annotated probe-lncRNA relationships was created. The official CLF file HuEx-1_0-st-v2.r2.clf was downloaded from the Affymetrix website (http://www.affymetrix.com/Auth/support/downloads/library_files/HuEx-1_0-st-v2.r2.zip).
Data analysis
Gene expression profiles were summarized by applying robust multi-array average (RMA)[18] normalization as implemented in affymetrix power tools (1.14.4 package apt-probeset-summarize), using the newly-created PGF file and the official CLF file.
The normalized data were analyzed with a Bioconductor package linear models for microarray data (LIMMA), a modified t-test incorporating the Benjamini-Hochberg multiple hypotheses correction technique[19]. Genes with adjusted P-values below 0.01 were considered differentially expressed. The heatmap of differentially expressed genes was generated using BRB-Array Tools Version 4.3.0 Beta 1 (http://linus.nci.nih.gov/BRB-ArrayTools.html)[20].
RESULTS
Re-annotation of exon array probes
A computational pipeline was established to re-annotate over 6.5 million probes of the Affymetrix Human Exon 1.0 ST array (Figure 1). There were 315255 probes perfectly matched to non-coding RNAs but not to any protein-coding transcript. These probes were mapped from transcript level to gene level and 278918 probes matched to one gene were retained. Probes mapping to short ncRNAs and pseudogenes were then discarded, leaving 136,533 probes mapping to lncRNAs, which were annotated with Ensembl (processed_transcript, lincRNA, antisense, non_coding, sense_intronic, ncrna_host, sense_overlapping and 3prime_overlapping_ncrna). To further increase accuracy, genes matched by less than three probes were discarded. Finally, we obtained 136053 probes uniquely mapping to lncRNAs at the gene level, corresponding to 9294 lncRNAs. The number of probes mapping to lncRNAs ranged from 3 to 257 and the average was 18.
Figure 1.
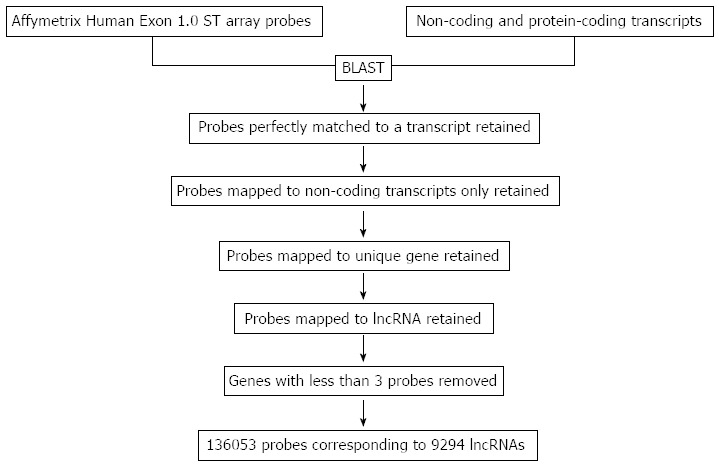
Computational pipeline for re-annotating the probes of the Affymetrix Human Exon 1.0 ST array. lncRNA: Long non-coding RNA.
Identification of differentially expressed lncRNAs in gastric cancer
The CEL files were processed by Affymetrix Power Tools for background correction, normalization, and summarizations with RMA algorithm. Using LIMMA with an adjusted P-value of less than 0.01 as a threshold, we identified 88 lncRNAs that were differentially expressed in gastric cancer as compared to normal gastric tissue (Figure 2). The top 30 lncRNAs differentially expressed in gastric cancer are listed in Table 1. Of 88 differentially expressed lncRNAs, 71 lncRNAs were found to be upregulated and 17 to be downregulated. Most of these lncRNAs do not have an official Human Genome Nomenclature Committee symbol and their function is unknown. But some have been reported to play a role in cancer, including LINC00152[21], taurine upregulated 1 (TUG1)[22], urothelial cancer associated 1 (UCA1)[23,24], Pvt1 oncogene (PVT1)[25], small nucleolar RNA host gene 1 (SNHG1)[26], and LINC00261[27].
Figure 2.
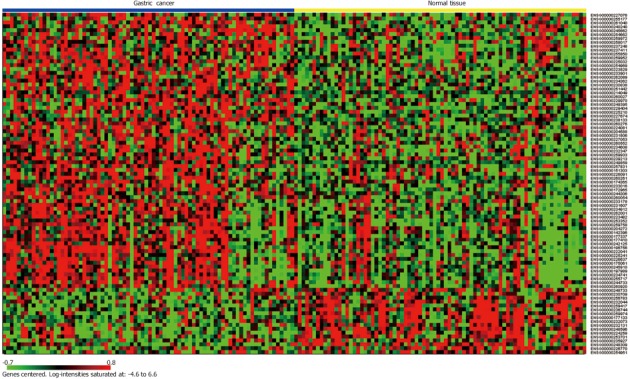
Clustering heatmap of 80 paired samples based on the 88 differentially expressed long non-coding RNAs. Each column represents one sample and each row represents one long non-coding RNA. Gene expression levels are indicated as follows: red, high expression; green, low expression.
Table 1.
Top 30 long non-coding RNAs differentially expressed in gastric cancer
| Ensembl gene ID | Fold change | Adjusted P-value | Gene biotype | HGNC symbol |
| ENSG00000222041 | 1.92678 | 6.89E-09 | lincRNA | LINC00152 |
| ENSG00000177410 | 1.54450 | 2.32E-06 | lincRNA | ZNFX1-AS1 |
| ENSG00000242125 | 1.65582 | 9.09E-06 | processed_transcript | SNHG3 |
| ENSG00000196756 | 1.31942 | 1.26E-05 | lincRNA | |
| ENSG00000255717 | 1.49039 | 2.30E-05 | ncrna_host | SNHG1 |
| ENSG00000226637 | 1.43434 | 2.30E-05 | lincRNA | |
| ENSG00000177337 | 1.40114 | 2.30E-05 | lincRNA | |
| ENSG00000249859 | 1.35920 | 2.98E-05 | lincRNA | PVT1 |
| ENSG00000197989 | 1.20284 | 6.10E-05 | lincRNA | SNHG12 |
| ENSG00000172965 | 1.23947 | 7.60E-05 | processed_transcript | |
| ENSG00000234741 | 1.37852 | 9.11E-05 | non_coding | GAS5 |
| ENSG00000177133 | 0.79465 | 9.11E-05 | processed_transcript | |
| ENSG00000255850 | 1.38946 | 1.45E-04 | antisense | |
| ENSG00000232131 | 0.69562 | 1.77E-04 | antisense | |
| ENSG00000244306 | 1.20990 | 2.64E-04 | lincRNA | |
| ENSG00000234608 | 1.20458 | 2.72E-04 | lincRNA | C12orf47 |
| ENSG00000259758 | 1.37499 | 2.72E-04 | antisense | |
| ENSG00000262001 | 1.24714 | 2.72E-04 | lincRNA | |
| ENSG00000259974 | 0.53383 | 3.56E-04 | lincRNA | LINC00261 |
| ENSG00000237248 | 1.32118 | 3.89E-04 | lincRNA | |
| ENSG00000249395 | 1.49799 | 4.04E-04 | lincRNA | |
| ENSG00000175061 | 1.21582 | 4.68E-04 | non_coding | C17orf76-AS1 |
| ENSG00000253352 | 1.39461 | 4.68E-04 | lincRNA | TUG1 |
| ENSG00000260920 | 1.46802 | 5.75E-04 | sense_overlapping | |
| ENSG00000248309 | 0.88939 | 5.94E-04 | lincRNA | |
| ENSG00000231806 | 1.36155 | 6.48E-04 | lincRNA | |
| ENSG00000223482 | 1.23001 | 6.48E-04 | antisense | |
| ENSG00000227076 | 1.22709 | 6.48E-04 | sense_intronic | |
| ENSG00000225241 | 1.20664 | 6.48E-04 | lincRNA | NBPF8 |
| ENSG00000236744 | 0.74935 | 6.48E-04 | processed_transcript |
HGNC: Human Genome Nomenclature Committee; TUG1: Taurine upregulated 1; SNHG: Small nucleolar RNA host gene.
Validation in an independent data set
To independently validate our results, we conducted the same analysis on GSE33335 and found that 59% of the differentially expressed lncRNAs identified by above analysis showed significant expression changes (adjusted P < 0.01) with the same direction. As shown in Figure 3, the distribution of expression differentials between the experimental data set and the validation data set is significantly concordant, reflecting a high consistence in expression patterns of these genes among different sample sets.
Figure 3.
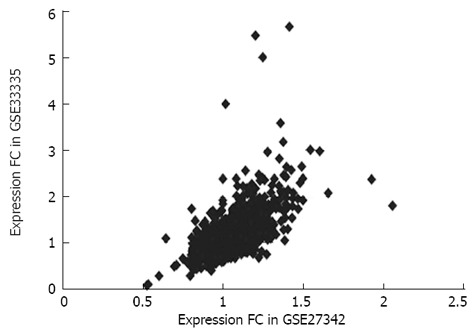
Distribution of expression differentials between experimental data set GSE27342 and validation data set GSE33335.
DISCUSSION
LncRNAs have comprehensive functions in biological processes through various mechanisms[6,7]. The expression patterns of lncRNAs are of great importance in the cancer field and are often investigated with tiling arrays[28,29], RNA sequencing[30] or lncRNA-specific microarrays[31,32], which are relatively expensive and inflexible. Recently, studies have suggested that lncRNA expression profiling may be achieved by mining existing microarray data because some probes uniquely mapping to lncRNAs are fortuitously represented on these arrays[33,34]. The Affymetrix Human Exon 1.0 ST array consists of over 6.5 million individual probes designed along the entire length of the gene as opposed to just the 3’ end, providing a unique platform for mining lncRNA profiles[35,36].
The lncRNA list used in this study was retrieved from Ensembl and is equivalent to the GENCODE lncRNA data set. This data set utilizes a combination of manual curation, computational analysis and targeted experimental approaches, and is the largest catalog of human lncRNAs to date[22]. To filter out potentially unrecognized probes mapping to protein-coding genes, we generated a merged known protein-coding gene list from RefSeq, UCSC and Ensembl. Still, some probes could potentially hybridize to other undiscovered transcripts or genes.
We identified 136053 probes from the Affymetrix Human Exon 1.0 ST array uniquely mapping to lncRNAs at the gene level. These probes correspond to 9294 lncRNAs, covering nearly 76% of the GENCODE lncRNA data set. This analysis revealed a set of lncRNAs that were differentially expressed between gastric cancer and normal gastric tissue, some of which have been previously reported in human cancers. For example, TUG1 is upregulated in bladder urothelial carcinoma and high TUG1 expression levels were associated with high grade and stage carcinomas[22]. Knockdown of TUG1 induced cell proliferation inhibition and apoptosis. Another candidate, UCA1, is dramatically upregulated in bladder cancer, suggesting it may be a very sensitive marker for bladder cancer[23]. Exogenous expression of UCA1 enhanced tumorigenicity, invasive potential, and drug resistance in BLS-211 cells[24]. Also, PVT1, located in 8q24 and amplified and overexpressed in ovarian and breast cancer, increases cell proliferation and inhibits apoptosis[25]. In light of published results in other cancers, we hypothesize that these lncRNAs may play an important role in the development of gastric cancer and are potential candidates for new biomarkers and therapeutic targets in gastric cancer.
Recently, H19 was shown to be upregulated in gastric cancer and its overexpression contributes to proliferation of gastric cancer cells[37]. In our study, the fold change of H19 in gastric cancer versus normal tissue is 1.378 with an adjusted P-value of 0.043. Though the difference was not statistically significant with our threshold (an adjusted P-value less than 0.001), the trend is consistent with the early report. However, our analysis may miss some lncRNAs that other groups have demonstrated to be involved in the development of gastric cancer due to the different distributions of the patient populations in terms of age, gender and cancer subtype and stage. We are also interested in exploring which lncRNAs are differentially expressed in different stages of gastric cancer. Unfortunately, the majority of gastric cancer samples in GSE27342 are stage III (54/72), making it is challenging to identify differentially expressed lncRNAs based on the stage of disease.
In conclusion, we presented global lncRNA expression profiles in gastric cancer by mining existing microarray data sets. We identified a set of lncRNAs that were differentially expressed in gastric cancer, revealing potential candidates for gastric cancer biomarkers, potentially improving diagnosis and therapy.
COMMENTS
Background
Long non-coding RNAs (lncRNAs) are an important class of regulatory transcripts involved in a variety of biological functions. While they are aberrantly expressed in many types of cancers, their expression patterns and biological functions in gastric cancer remain unknown.
Research frontiers
LncRNA expression profiles are often investigated with tiling arrays, RNA sequencing or lncRNA-specific microarrays. Existing microarray data represent unique probes specific to lncRNAs, suggesting lncRNA expression profiling may be achieved by mining existing microarray data.
Innovations and breakthroughs
The authors re-annotated the probes of the Affymetrix Human Exon 1.0 ST array and identified probes uniquely mapping to lncRNAs at the gene level. By analyzing a publicly available data set, a set of lncRNAs differentially expressed in gastric cancer were identified.
Applications
The study results suggest lncRNAs play an important role in the development of gastric cancer and have the potential to be used as molecular diagnostic markers and therapeutic targets in gastric cancer.
Terminology
LncRNAs are non-protein coding transcripts having a size greater than 200 nucleotides. This limit distinguishes lncRNAs from small non-coding RNAs such as microRNAs, short interfering RNAs, Piwi-interacting RNAs, and small nucleolar RNAs.
Peer review
The authors investigated the expression patterns of lncRNAs in gastric cancer by mining existing microarray data sets. A set of lncRNAs differentially expressed in gastric cancer were identified. These results are interesting and suggest that lncRNAs may play an important role in gastric cancer.
Footnotes
P- Reviewers Chen WX, Mann O, Martin-Villa JM S- Editor Zhai HH L- Editor A E- Editor Zhang DN
References
- 1.Kapranov P, Cawley SE, Drenkow J, Bekiranov S, Strausberg RL, Fodor SP, Gingeras TR. Large-scale transcriptional activity in chromosomes 21 and 22. Science. 2002;296:916–919. doi: 10.1126/science.1068597. [DOI] [PubMed] [Google Scholar]
- 2.Kapranov P, Willingham AT, Gingeras TR. Genome-wide transcription and the implications for genomic organization. Nat Rev Genet. 2007;8:413–423. doi: 10.1038/nrg2083. [DOI] [PubMed] [Google Scholar]
- 3.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 6.Nagano T, Fraser P. No-nonsense functions for long noncoding RNAs. Cell. 2011;145:178–181. doi: 10.1016/j.cell.2011.03.014. [DOI] [PubMed] [Google Scholar]
- 7.Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482:339–346. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 9.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cui J, Chen Y, Chou WC, Sun L, Chen L, Suo J, Ni Z, Zhang M, Kong X, Hoffman LL, et al. An integrated transcriptomic and computational analysis for biomarker identification in gastric cancer. Nucleic Acids Res. 2011;39:1197–1207. doi: 10.1093/nar/gkq960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cui J, Li F, Wang G, Fang X, Puett JD, Xu Y. Gene-expression signatures can distinguish gastric cancer grades and stages. PLoS One. 2011;6:e17819. doi: 10.1371/journal.pone.0017819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng L, Wang P, Yang S, Yang Y, Zhang Q, Zhang W, Xiao H, Gao H, Zhang Q. Identification of genes with a correlation between copy number and expression in gastric cancer. BMC Med Genomics. 2012;5:14. doi: 10.1186/1755-8794-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng L, Yang S, Yang Y, Zhang W, Xiao H, Gao H, Deng X, Zhang Q. Global gene expression and functional network analysis of gastric cancer identify extended pathway maps and GPRC5A as a potential biomarker. Cancer Lett. 2012;326:105–113. doi: 10.1016/j.canlet.2012.07.031. [DOI] [PubMed] [Google Scholar]
- 14.Flicek P, Amode MR, Barrell D, Beal K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fairley S, Fitzgerald S, et al. Ensembl 2012. Nucleic Acids Res. 2012;40:D84–D90. doi: 10.1093/nar/gkr991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dreszer TR, Karolchik D, Zweig AS, Hinrichs AS, Raney BJ, Kuhn RM, Meyer LR, Wong M, Sloan CA, Rosenbloom KR, et al. The UCSC Genome Browser database: extensions and updates 2011. Nucleic Acids Res. 2012;40:D918–D923. doi: 10.1093/nar/gkr1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pruitt KD, Tatusova T, Brown GR, Maglott DR. NCBI Reference Sequences (RefSeq): current status, new features and genome annotation policy. Nucleic Acids Res. 2012;40:D130–D135. doi: 10.1093/nar/gkr1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 19.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 20.Simon R, Lam A, Li MC, Ngan M, Menenzes S, Zhao Y. Analysis of gene expression data using BRB-ArrayTools. Cancer Inform. 2007;3:11–17. [PMC free article] [PubMed] [Google Scholar]
- 21.Neumann O, Kesselmeier M, Geffers R, Pellegrino R, Radlwimmer B, Hoffmann K, Ehemann V, Schemmer P, Schirmacher P, Lorenzo Bermejo J, et al. Methylome analysis and integrative profiling of human HCCs identify novel protumorigenic factors. Hepatology. 2012;56:1817–1827. doi: 10.1002/hep.25870. [DOI] [PubMed] [Google Scholar]
- 22.Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa A, Searle S, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang XS, Zhang Z, Wang HC, Cai JL, Xu QW, Li MQ, Chen YC, Qian XP, Lu TJ, Yu LZ, et al. Rapid identification of UCA1 as a very sensitive and specific unique marker for human bladder carcinoma. Clin Cancer Res. 2006;12:4851–4858. doi: 10.1158/1078-0432.CCR-06-0134. [DOI] [PubMed] [Google Scholar]
- 24.Wang F, Li X, Xie X, Zhao L, Chen W. UCA1, a non-protein-coding RNA up-regulated in bladder carcinoma and embryo, influencing cell growth and promoting invasion. FEBS Lett. 2008;582:1919–1927. doi: 10.1016/j.febslet.2008.05.012. [DOI] [PubMed] [Google Scholar]
- 25.Guan Y, Kuo WL, Stilwell JL, Takano H, Lapuk AV, Fridlyand J, Mao JH, Yu M, Miller MA, Santos JL, et al. Amplification of PVT1 contributes to the pathophysiology of ovarian and breast cancer. Clin Cancer Res. 2007;13:5745–5755. doi: 10.1158/1078-0432.CCR-06-2882. [DOI] [PubMed] [Google Scholar]
- 26.Berretta R, Moscato P. Cancer biomarker discovery: the entropic hallmark. PLoS One. 2010;5:e12262. doi: 10.1371/journal.pone.0012262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin ZY, Chuang WL. Genes responsible for the characteristics of primary cultured invasive phenotype hepatocellular carcinoma cells. Biomed Pharmacother. 2012;66:454–458. doi: 10.1016/j.biopha.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 28.Perez DS, Hoage TR, Pritchett JR, Ducharme-Smith AL, Halling ML, Ganapathiraju SC, Streng PS, Smith DI. Long, abundantly expressed non-coding transcripts are altered in cancer. Hum Mol Genet. 2008;17:642–655. doi: 10.1093/hmg/ddm336. [DOI] [PubMed] [Google Scholar]
- 29.Silva JM, Perez DS, Pritchett JR, Halling ML, Tang H, Smith DI. Identification of long stress-induced non-coding transcripts that have altered expression in cancer. Genomics. 2010;95:355–362. doi: 10.1016/j.ygeno.2010.02.009. [DOI] [PubMed] [Google Scholar]
- 30.Prensner JR, Iyer MK, Balbin OA, Dhanasekaran SM, Cao Q, Brenner JC, Laxman B, Asangani IA, Grasso CS, Kominsky HD, et al. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat Biotechnol. 2011;29:742–749. doi: 10.1038/nbt.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang F, Zhang L, Huo XS, Yuan JH, Xu D, Yuan SX, Zhu N, Zhou WP, Yang GS, Wang YZ, et al. Long noncoding RNA high expression in hepatocellular carcinoma facilitates tumor growth through enhancer of zeste homolog 2 in humans. Hepatology. 2011;54:1679–1689. doi: 10.1002/hep.24563. [DOI] [PubMed] [Google Scholar]
- 32.Yu G, Yao W, Wang J, Ma X, Xiao W, Li H, Xia D, Yang Y, Deng K, Xiao H, et al. LncRNAs expression signatures of renal clear cell carcinoma revealed by microarray. PLoS One. 2012;7:e42377. doi: 10.1371/journal.pone.0042377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Michelhaugh SK, Lipovich L, Blythe J, Jia H, Kapatos G, Bannon MJ. Mining Affymetrix microarray data for long non-coding RNAs: altered expression in the nucleus accumbens of heroin abusers. J Neurochem. 2011;116:459–466. doi: 10.1111/j.1471-4159.2010.07126.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liao Q, Liu C, Yuan X, Kang S, Miao R, Xiao H, Zhao G, Luo H, Bu D, Zhao H, et al. Large-scale prediction of long non-coding RNA functions in a coding-non-coding gene co-expression network. Nucleic Acids Res. 2011;39:3864–3878. doi: 10.1093/nar/gkq1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Okoniewski MJ, Yates T, Dibben S, Miller CJ. An annotation infrastructure for the analysis and interpretation of Affymetrix exon array data. Genome Biol. 2007;8:R79. doi: 10.1186/gb-2007-8-5-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gardina PJ, Clark TA, Shimada B, Staples MK, Yang Q, Veitch J, Schweitzer A, Awad T, Sugnet C, Dee S, et al. Alternative splicing and differential gene expression in colon cancer detected by a whole genome exon array. BMC Genomics. 2006;7:325. doi: 10.1186/1471-2164-7-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang F, Bi J, Xue X, Zheng L, Zhi K, Hua J, Fang G. Up-regulated long non-coding RNA H19 contributes to proliferation of gastric cancer cells. FEBS J. 2012;279:3159–3165. doi: 10.1111/j.1742-4658.2012.08694.x. [DOI] [PubMed] [Google Scholar]


