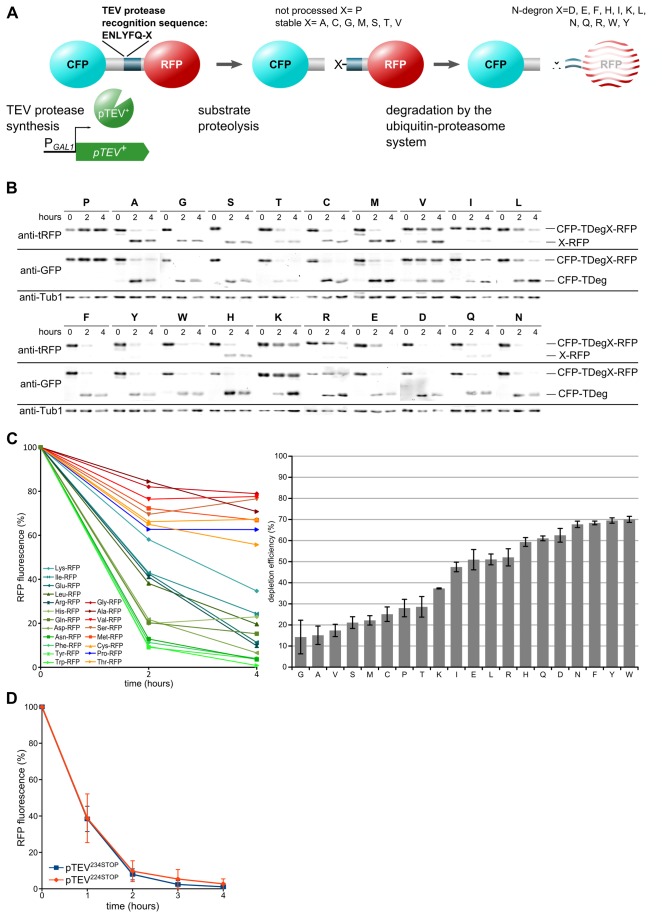
Figure 1. Activation of the N-degron is rate limiting during substrate depletion by the TIPI system.
A) TIPI efficiency is influenced by three factors, synthesis of the TEV protease by the galactose-inducible GAL1 promoter, proteolysis of the recognition sequence, and degradation of the target protein by the ubiquitin-proteasome system. A reporter protein consisting of two fluorescent proteins (cyan and red) fused together by the TDegX sequence containing the TEV protease recognition sequence (X= amino acid at position P1') and the N-degron sequence. Please note that we follow the original classification of stabilizing and destabilizing residues without considering N-degrons that are produced by N-acetylation. In our constructs, histidine follows X, which prevents acetylation of X in case of Met, Ala, Ser, Cys, Thr, and Val. B) In vivo analysis of the P1' specificity of the pTEV+ protease. Processing of the tester constructs CFP-TDegX-RFP (plasmid based) was observed after induction of pTEV+ protease production (PGAL1 -pTEV + in strain YCT1169) by addition of galactose (2% final concentration). Total cell extracts were fractionated by SDS-polyacrylamide electrophoresis, followed by immunoblotting with antibodies directed against GFP, tRFP and Tub1 (loading control). C) Quantification of X-RFP depletion. RFP fluorescence (same constructs as in B) was measured by a fluorimeter after induction of pTEV+ protease synthesis (left graph) and the depletion efficiency of the different substrates was calculated (right graph). Curves are mean values of at least four measurements, normalized to initial RFP fluorescence. Depletion efficiency is represented by the area above each curve (error bars: SEM). D) C-terminal truncation of the TEV protease at position 224 does not influence its activity. The abundance of the tester substrate CFP-TDegF-RFP was followed over time after expression of different pTEV protease variants by fluorimeter measurements (conditions as in C). The plasmid pDS7 was used to express the substrate in yeast strains YCT1243 and YCT1244; error bars represent standard deviation; each construct was measured at least five times.