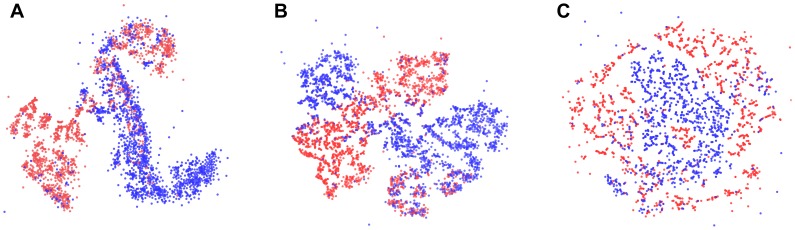
Figure 5. Data distribution in the learned feature spaces suggests disease subpopulations.
A: First-layer features. B: Second-layer features. C: Expert engineered features. These two-dimensional embeddings using t-SNE suggest several subpopulations of gout (red) and leukemia (blue) in both learned feature spaces. We suspect that these subpopulations largely represent differences in treatment approach, but they may also be illuminating pathophysiologic differences. The engineered feature space separates the two known phenotypes adequately for a discrimination task, but offers only weak suggestions of subpopulations: without the colors corresponding to known phenotypes, it would be difficult to identify more than a single large cluster in this space. The t-SNE algorithm preserves near neighbor distances at the expense of far neighbor distances, so we cannot draw conclusions from the macro-scale shape or relative orientation of the clusters, only their number and substructure.