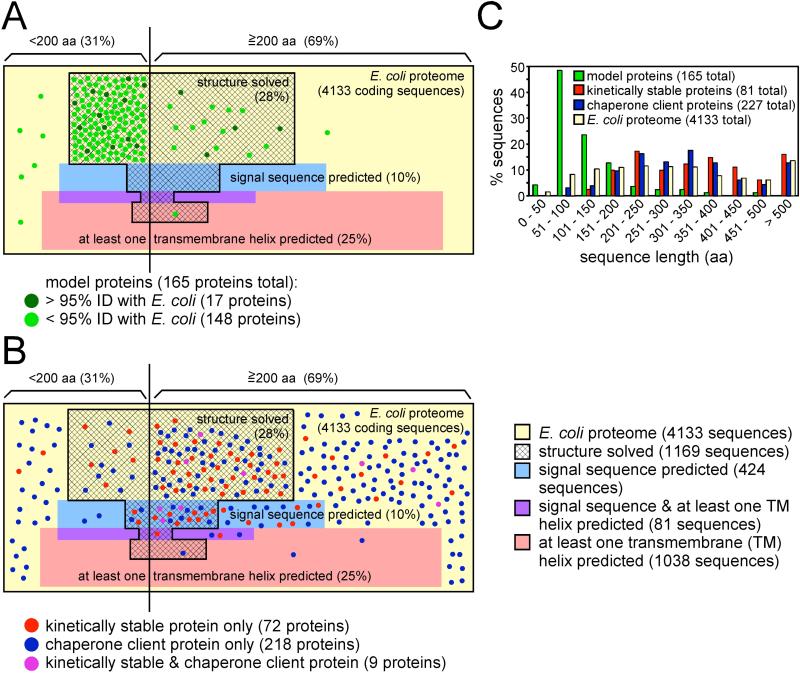
Figure 1. While typical protein folding models exhibit properties not representative of the E. coli proteome, emerging techniques can capture a broader set of proteins.
(A) The 4133 proteins from E. coli str. K-12 substr. MG1655 (NC_000913.2) were used to construct a proportional Venn diagram, with each unit area in the yellow rectangle corresponding to one E. coli protein coding sequence. These sequences were divided by length (< or ≥200 aa) and analyzed for the presence of an N-terminal signal sequence (http://www.cbs.dtu.dk/services/SignalP) (blue shading), one or more transmembrane α-helices (http://www.cbs.dtu.dk/services/TMHMM) (pink shading), both a signal sequence and a transmembrane α-helix (purple shading) and/or a PDB entry with >95% sequence identity to at least some portion of the protein sequence (hatched area). Note that this map underestimates the complexity of the proteome, as each protein coding sequence from E. coli genome is treated as a separate monomeric protein. A set of 165 non-redundant model proteins used to study protein folding (<95% sequence identity) [3-9] was also analyzed. Each protein is indicated by a green point proportional to the size of one E. coli coding sequence. Seventeen of the model proteins have >95% sequence identity to an E. coli protein (dark green points); the remaining 148 model proteins are from other organisms (light green points). In some cases these models represent individual domains or fragments taken from larger proteins, but as it is known that removal from a larger protein context can change folding behavior [33, 44] (see text), the size of the studied domain is used here. (B) Subsets of proteins identified by proteome-wide screens designed to select other, non-traditional folding behavior were categorized as described for the 165 folding models and compared to the properties of the E. coli proteome as in panel (A). Kinetically stable proteins (red points) were identified by protease resistance [43] or resistance to moderate concentrations of sodium dodecyl sulfate (SDS) [42], yielding 81 non-redundant E. coli proteins. E. coli chaperone client proteins (blue points) represent both DnaK substrates (category “enriched” in [61]) and GroEL substrates (“class IV” in [60]), resulting in a set of 227 proteins. Proteins present in both sets (kinetically stable and chaperone client) are indicated as purple points. Note that there is only one protein in common between the folding models (panel (A)) and kinetically stable and/or chaperone client proteins: maltose binding protein, a kinetically stable protein [43]. (C) Size distribution for each protein group shown in panels (A) and (B), sorted by sequence length.