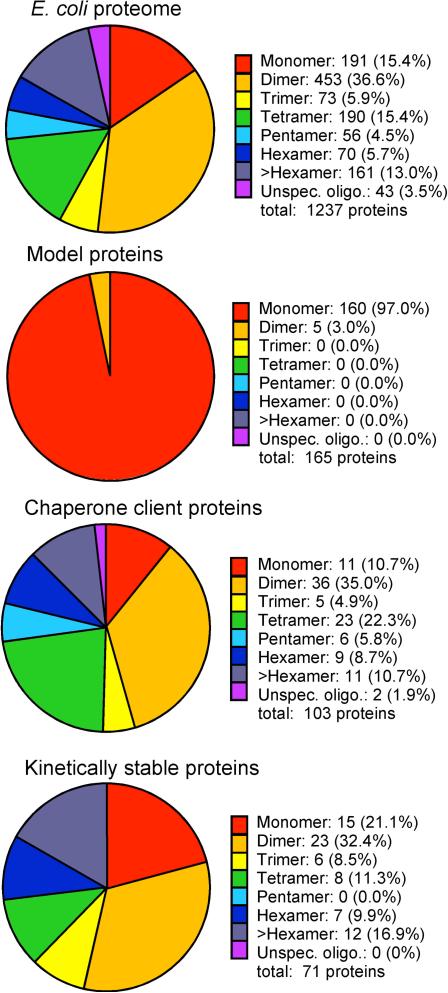
Figure 2. Protein folding models are biased towards monomeric proteins.
The multimerization state of each group of proteins shown in Figure 1 (E. coli proteome, protein folding models, kinetically stable proteins, chaperone client proteins) was determined. For the E. coli proteome, subunit assignments in the Uniprot database were used (30% of proteins in the E. coli proteome have assignments; 1236 proteins). Multimerization state for the 165 non-redundant protein folding models was assigned based on reported multimerization state in the protein folding literature. The multimerization state is indicated for 71 of the 81 kinetically stable proteins identified in ref. [42, 43]. The multimerization state of the chaperone client proteins was assigned using the Uniprot database. 103 of the 227 non-redundant chaperone client proteins have a subunit assignment in the Uniprot database.