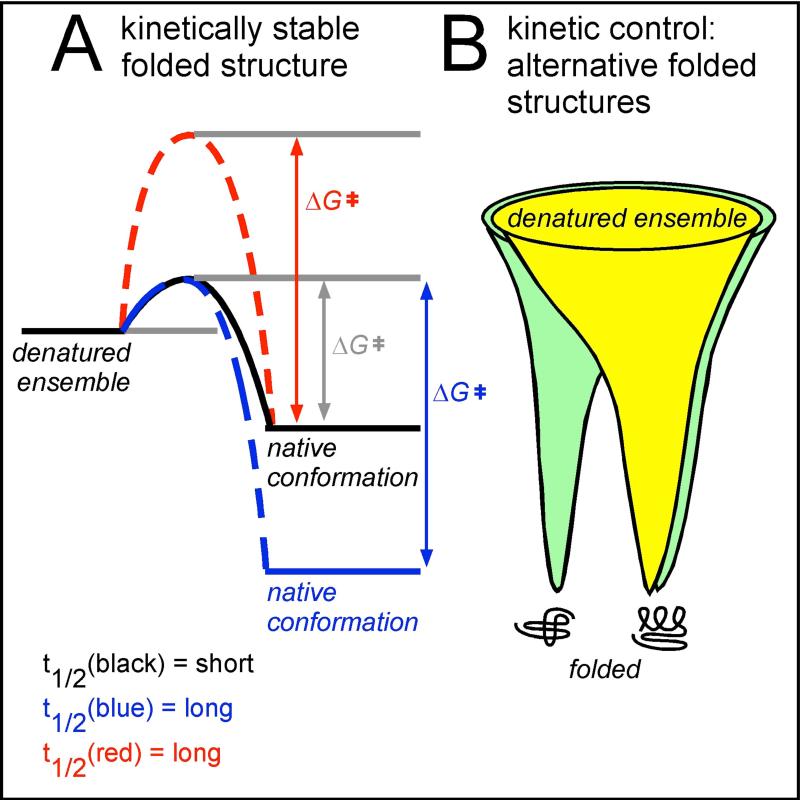
Figure 3. Examples of diversity amongst protein folding mechanisms.
(A): Most proteins currently used as folding models are marginally stable (black), meaning that their folded lifetime (t1/2) is short. Lifetime can be increased in two ways. The native structure can be stabilized thermodynamically, increasing the energetic difference between the denatured ensemble and the native structure (increasing ΔGofolding, blue). Alternatively, the energetic barrier separating the denatured ensemble and the native conformation (ΔG‡) can be increased (red); this will preserve the (low) thermodynamic stability but increase the folded state lifetime. Increasing the energy barrier yields kinetically stable proteins, which can be identified by proteome-wide folding screens [42, 43] (see also Figure 1B). (B): Proteins fold from an ensemble of unfolded states, represented by the wide top of a protein folding funnel. In simple model systems (yellow), the funnel has one energy minimum, the native conformation. However, some proteins have a more complex energy landscape and can adopt alternative folded structures (green). These two folded structures may interconvert, or features of the cellular environment may stabilize a subset of early folding intermediates, resulting in a biased accumulation of one structure versus the other(s).