Figure 2. Techniques used for identification of chromosomal translocations and characterization of nuclear organization.
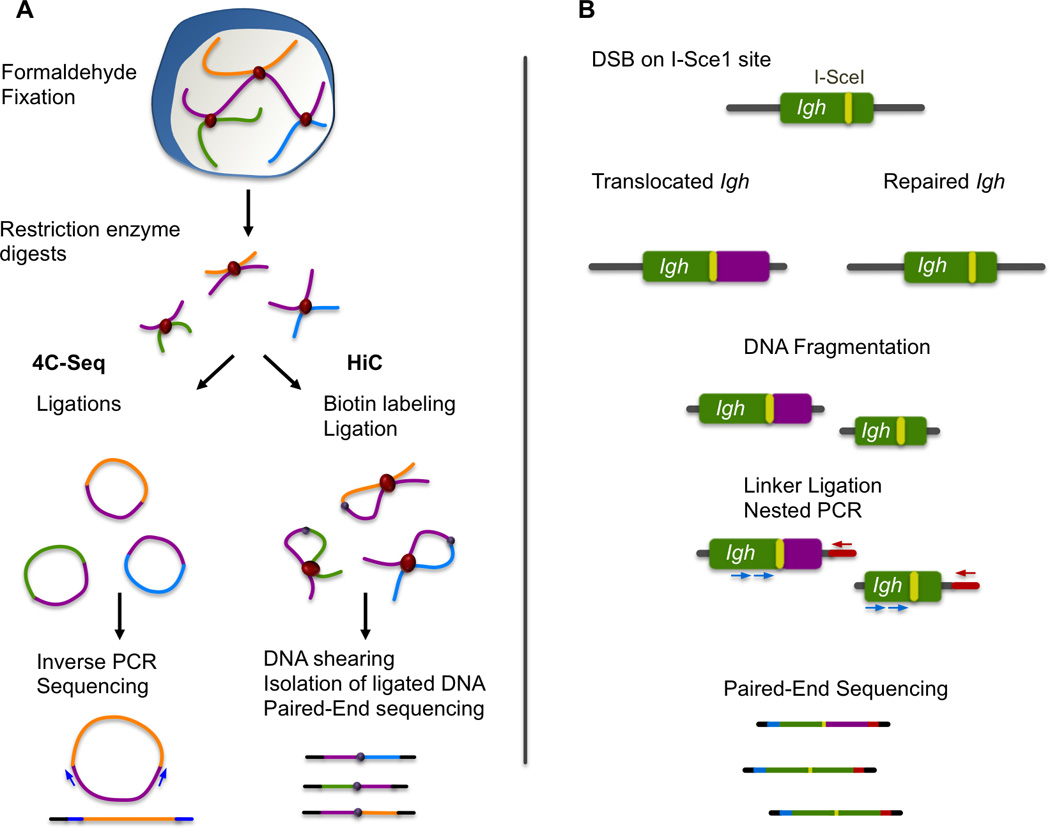
(A) 4C-seq and HiC identify chromosomal interactions through formaldehyde fixation and subsequent digestion and ligation of chromatin segments found in close proximity in the cell nucleus. 4C-seq is used to find all the interaction partners of a certain locus through inverse PCR and massively parallel sequencing. An additional digestion and ligation step (not represented here for simplification purposes) is required in this method. In HiC, all fragments that represent chromosomal interactions are isolated through biotin/streptavidin purification and then identified through genomewide sequencing. (B) Simplified scheme of one of the techniques used for genome-wide identification of Igh translocation partners. An engineered I-SceI site on Igh is used to induce breaks and translocations. These are identified through ligation-mediated PCR that takes advantage of the fact that the I-SceI site provides the precise location of one of the translocation partners.
