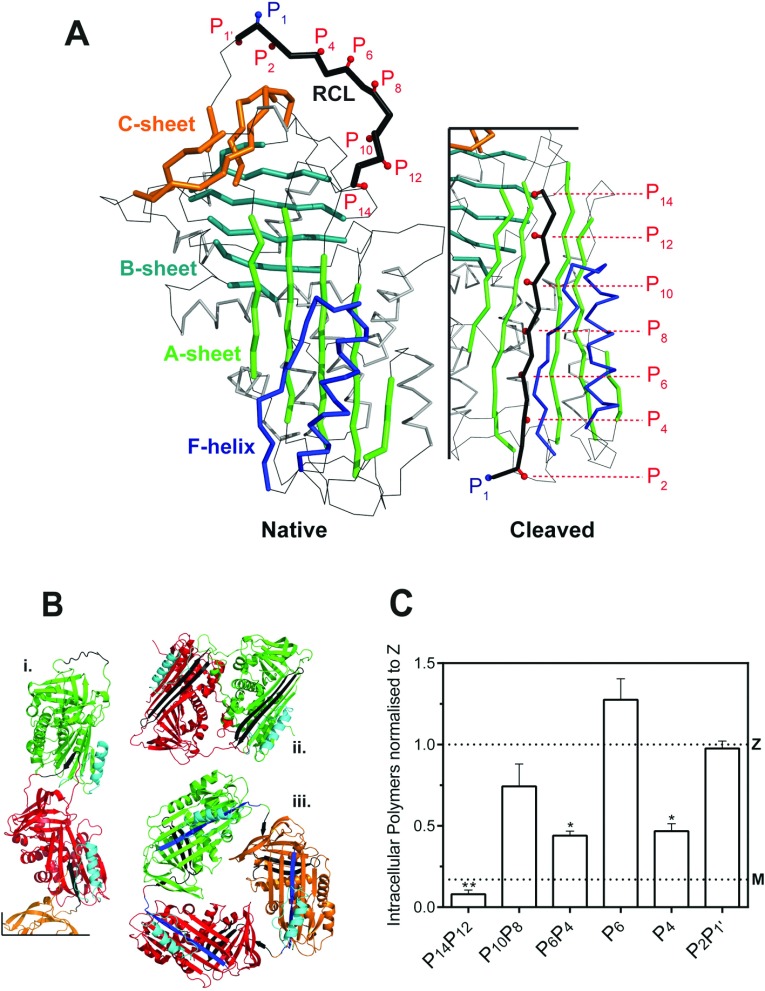
Figure 1. RCL mutants of α1-antitrypsin.
(A) The location of the mutations used in this study are indicated against a cartoon representation of wild-type α1-antitrypsin (prepared using PyMol and PDB entries 1QLP [42] and 1EZX [43]). The RCL is highlighted in black, and numbered according to the P-site convention of Schechter and Berger [28], in which the P1 and P1′ positions are either side of the site of cleavage by a cognate protease. The three β-sheets of the serpin fold and the F-helix are labelled. Upon transition from the native, active conformation (left panel) to the cleaved or latent forms, the RCL moves from an exposed to an inserted position as an additional β-strand of β-sheet A (right panel). (B) The three main models of polymerization: i, a molecular model of a loop-sheet polymer; ii, the crystal structure of a closed dimer of antithrombin; iii, a closed trimer of α1-antitrypsin. The site of inter-molecular interaction in each form is shown in black and helix F is coloured cyan. (C) Mutations at positions P14P12, P6P4 and P4 of the RCL reduce intracellular polymerization of Z α1-antitrypsin. COS-7 cells were transiently transfected with M α1-antitrypsin, Z α1-antitrypsin and RCL mutants on a Z α1-antitrypsin background in serum-free medium for 24 h before lysis. A sandwich ELISA analysis of intracellular polymers made use of the polymer-specific 2C1 monoclonal antibody. The results are normalized to Z α1-antitrypsin (n=4; mean±S.D.) and differences were assessed by one-way ANOVA with Bonferroni's correction for multiple comparisons: *, P<0.01; **, P<0.001. The value for M α1-antitrypsin is shown with a dotted line (M).