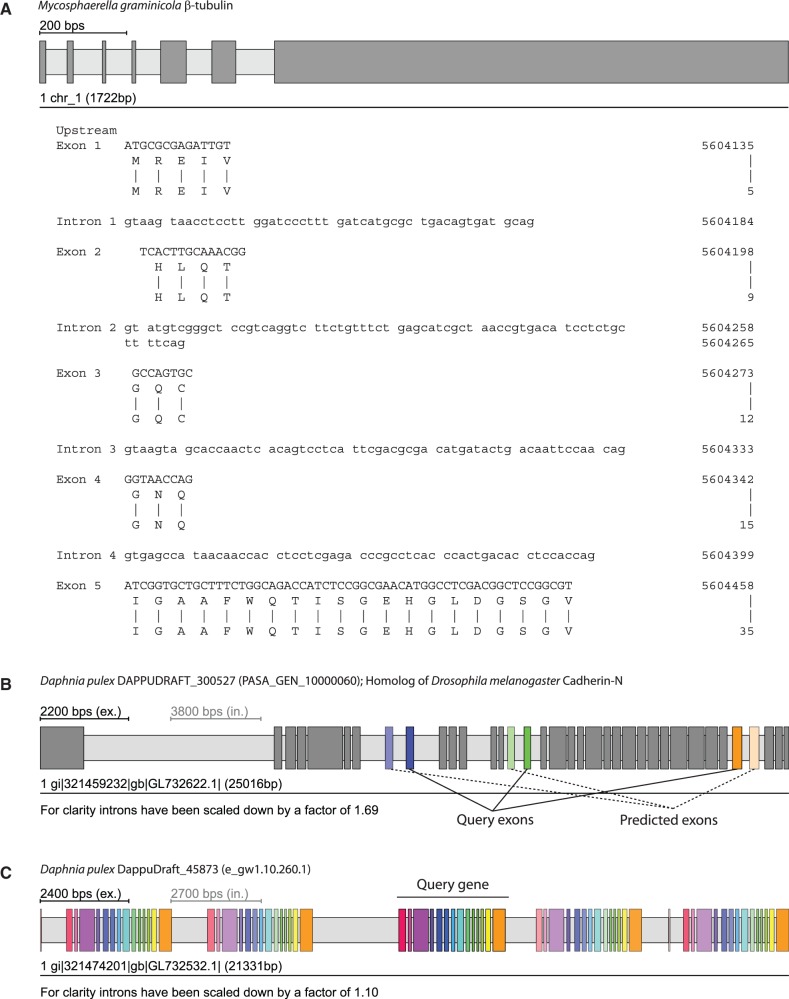
Figure 2.
Examples for running WebScipio using previously implemented advanced options and prediction algorithms. Exons are represented as dark-grey or coloured bars, and introns as light-grey bars. (A) The N-termini of the β-tubulin genes in fungi (here Mycosphaerella graminicola) are encoded by several very short exons. These exons are not recognized by ab initio gene prediction programs and hardly identified in other gene reconstruction software. (B) Predicted clusters of MXEs are displayed in colour. The exon of the cluster, which was present in the query sequence, is shown in full colour, whereas the predicted MXEs are shown with opacity that correlates to their similarity to the query exon (high similarity = high opacity). (C) Tandemly arrayed gene duplicates can be predicted within a user, given genomic region. Here, the query gene was the central gene of the cluster. In general, homologous exons in duplicated genes have the same colour as the exons of the query gene. Like with the MXE colouring, similarity to the query exon is expressed through opacity. Exceptions are exons whose homologues are fused or split exons in the duplicated gene.