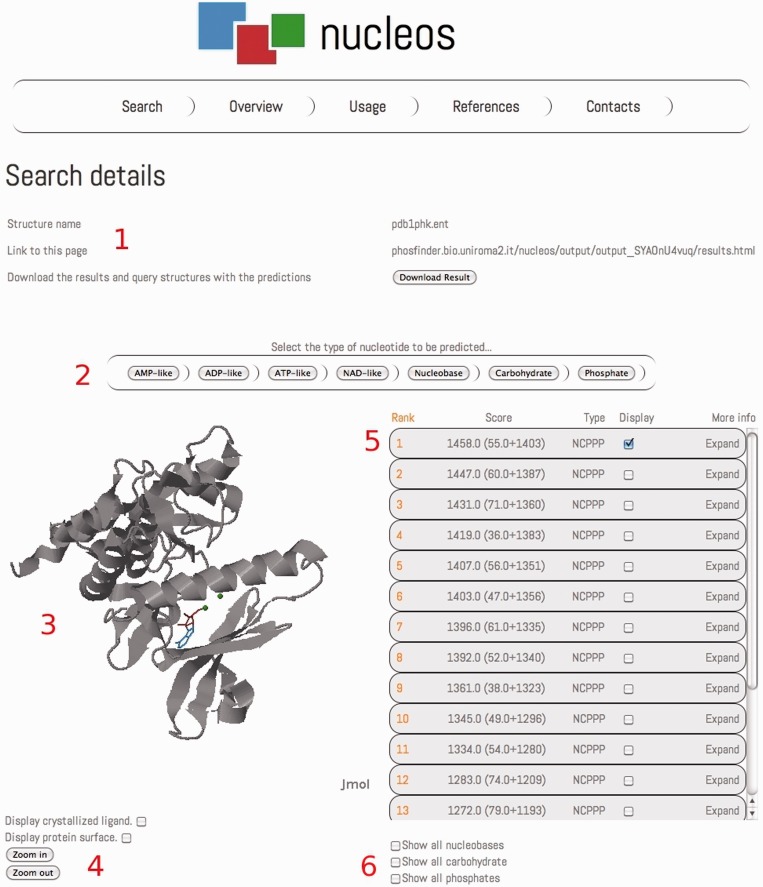
Figure 1.
An example of the Nucleos output page. The details of the query are displayed on the top of the page (1), reporting the name of the structure, the address of the results page and a download link to get a folder containing results in form of table and of PDB structures containing the predictions. A menu is provided to quickly navigate the results, switching among the different nucleotide predictions (2). A Jmol applet displays the query protein structure with the prediction (3), when switching to results for a different nucleotide type, the first prediction in the ranking is displayed. Buttons are provided to interact with the Jmol applet (4): zoom in/out, show/hide protein surface and crystallized ligand. Results for ATP-like predictions are ranked in the results table on the right (5). Buttons are provided to quickly show/hide all the predicted nucleobase, carbohydrate and phosphate binding sites (6).