Figure 2.
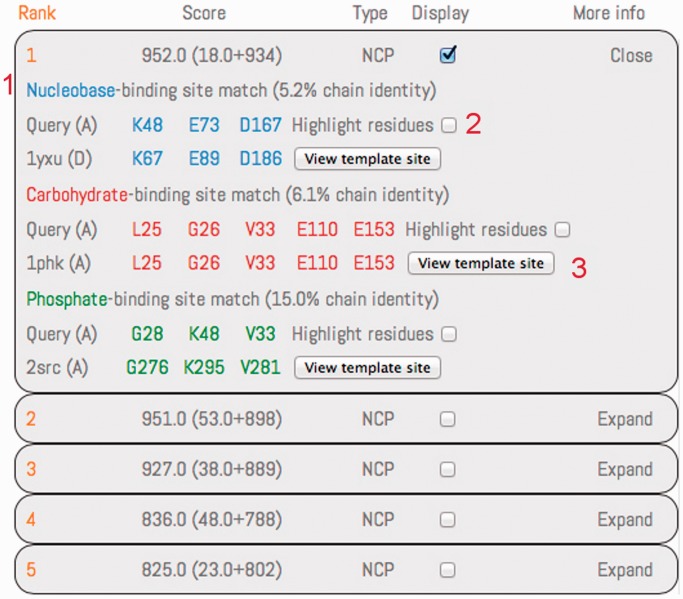
The results section for the first ranked prediction has been expanded using the button on the top right. Predictions are ranked by their score, with the scoring components reported within brackets (clustering score plus conservation score). The type of the prediction is reported using the ‘N’, ‘C’ and ‘P’ letters for the nucleobase, carbohydrate and phosphate, respectively. Results are displayed for all the modules composing the prediction, following a color code (blue for nucleobase, red for the carbohydrate and green for the phosphate). The structural similarity between residues of the query protein and a template binding site is detailed with the involved residues, paired following the structural match. The sequence identity, between the query protein structure and the protein containing the template-binding match involved in the similarity, is reported for each module. A checkbox is provided to highlight/bleach the residue in the Jmol applet. The template-binding site, highlighted in its original PDB structure, can be displayed in a popup Jmol applet.