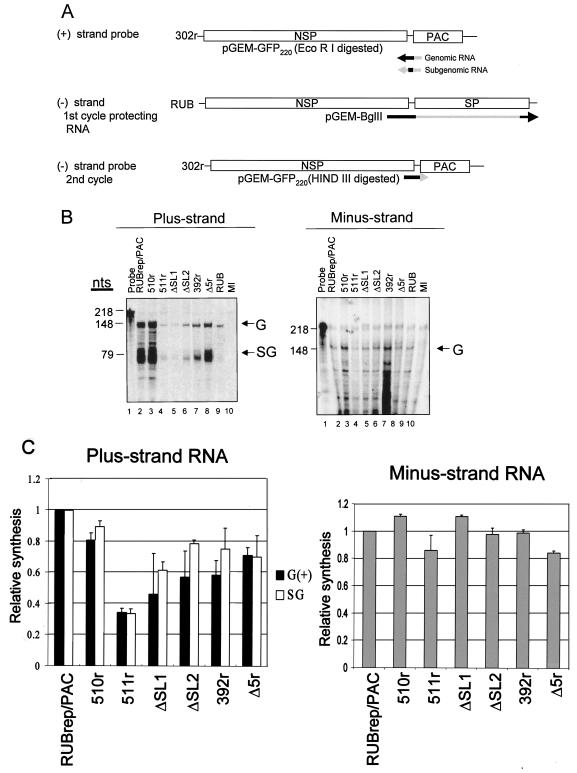
FIG. 4.
Strand-specific RNA synthesis in BHK cells transfected with various RUBrep/PAC constructs. (A) Schematic representation of probes and protecting RNA used for RPA. To detect plus-strand RNAs, including genomic and subgenomic RNAs, 35S-labeled RNA transcripts of negative polarity with a total of 218 nt were prepared by runoff transcription in vitro using digested pGEM-GFP220 and were used directly in RPA. To detect minus-strand RNA, unlabeled RNA probe transcribed from a construct, pGEM-BglII, encoding 3′ 4,000 nt in the RUB genome, was first used, and the protected products from this first cycle of RPA were hybridized to a 35S-labeled pGEM-GFP220 probe of positive polarity. The regions that would hybridize to replicon genomes are denoted as black lines, while those that would be digested from protecting RNA and probes by RNase are shown in gray. Equal amounts of RNA extracted from cells within five passages following puromycin were used for RPA analysis. (B) Representative gels are shown: left panel, plus strand RNA; right panel, minus strand RNA. The product protected by genomic (G) RNA (both plus and minus strand) was 148 nt, and that protected by the subgenomic RNA (SG) was 79 nt, as indicated on the right margin of the gel. RPA band intensities were quantitated using a FluorChem imaging system. (C) The results of quantitation are the means of three RPA analyses using intracellular RNA from two independent transfections.