Figure 1.
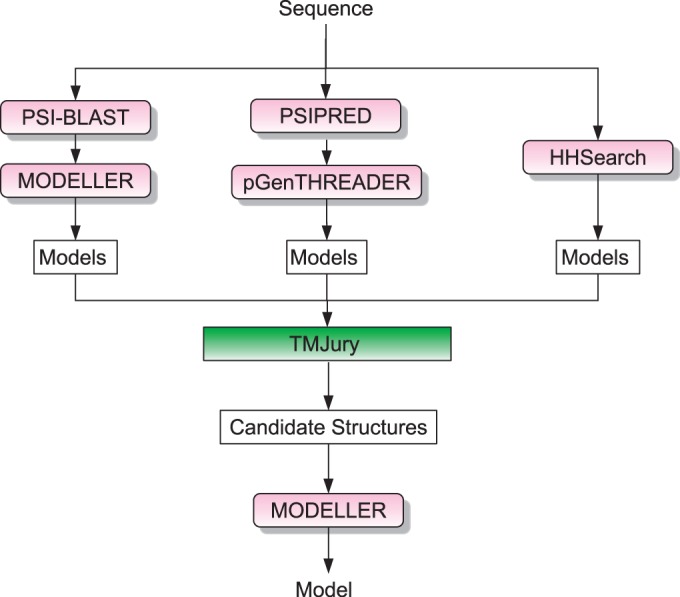
Flowchart of the BioSerf2 automated homology modelling protocol. Incoming query sequences are independently matched to PDB chains using PSIBLAST, pGenTHREADER and HH. The three sets of models produced are then compared by the TMJury process, which produces up to 10 candidate homologous structures. These structures and their alignments to the query sequence are used as input for MODELLER to produce a single final structure.
