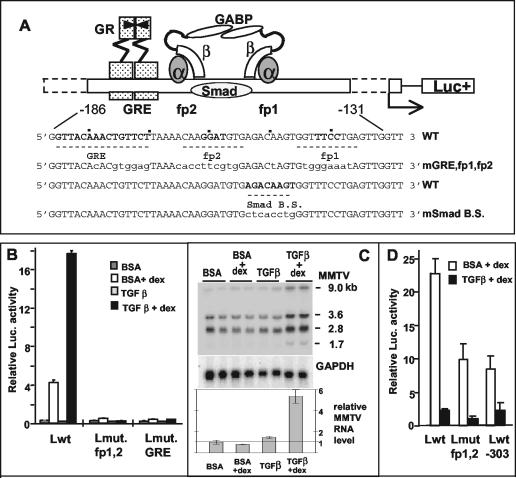
FIG. 1.
Opposite effects of TGF-β1 in a B-lymphoma cell line (B and C) versus a mammary tumor cell line (D). (A) Scheme of the arrangement of factors binding to the regulatory elements upstream of the MMTV promoter that are relevant for the present study. In the wild-type sequence (WT), the numbering refers to the transcription initiation site, and fp1 and fp2 denote the DNase I footprints with M12-cell nuclear extracts (5), with the core sequences for GABP binding in boldface. The putative Smad binding site is also marked. Mutated bases in the different reporter constructs are in lowercase letters (mGRE, fp1, and fp2 are mutated in all three sites; mSmad B.S. is mutated in the putative Smad binding site [modified from references 27 and 48]). (B and D) pGL-3 plasmids (B, 3 μg; D, 2 μg) with various LTRs were transiently transfected into M12 B cells (B) or GR mouse mammary tumor cells (D) that were kept thereafter in serum-free medium plus 5 ng of TGF-β1/ml or the carrier solution containing bovine serum albumin for 24 h (B) or 46 h (D). The “+dex” samples received Dex (50 [B] or 300 [D] nM) for the last 4 h (B) or 8 h (D) before cell lysis for determination of luciferase activity. The LTRs used were as follows: wild type (Lwt), LTR mutated in the fp1/fp2 GABP-binding sites (Lmut fp1,2), LTR mutated in the GRE (Lmut. GRE [LS−193/−162] [10]), and LTR 5′ truncated at position −303 (Lwt-303). The values for firefly luciferase activity were normalized against the Renilla luciferase activity produced by cotransfected plasmids (50 ng of pRL-SV40Δ enhancer [B] or 200 ng of pRL-SV40 [D]). Error bars denote the standard deviations in duplicate samples. The experiment in panel D was performed twice; the experiment in panel B was performed many times. (C) Cooperative effect of TGF-β1 and Dex on the endogenous MMTV RNA levels of M12 cells. Northern blot analysis of total RNA from M12 cells treated with TGF-β1 (5 ng/ml) or bovine serum albumin buffer for 24 h in serum-free medium in the presence or absence of 50 nM Dex for the last 4 h before RNA extraction. Total RNA was separated in a 1% agarose gel containing formaldehyde and blotted onto a nylon membrane, which was hybridized with a 32P-labeled probe of the MMTV env region (upper panel) and then with a probe of the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) cDNA as a loading control (lower panel). RNA samples are in duplicate. On the right, the sizes in kilobases of the viral mRNAs are given (see the text). The total intensity of the MMTV bands relative to the GAPDH control was quantified and plotted (relative MMTV RNA; values ± the standard deviation are indicated).