FIG. 2.
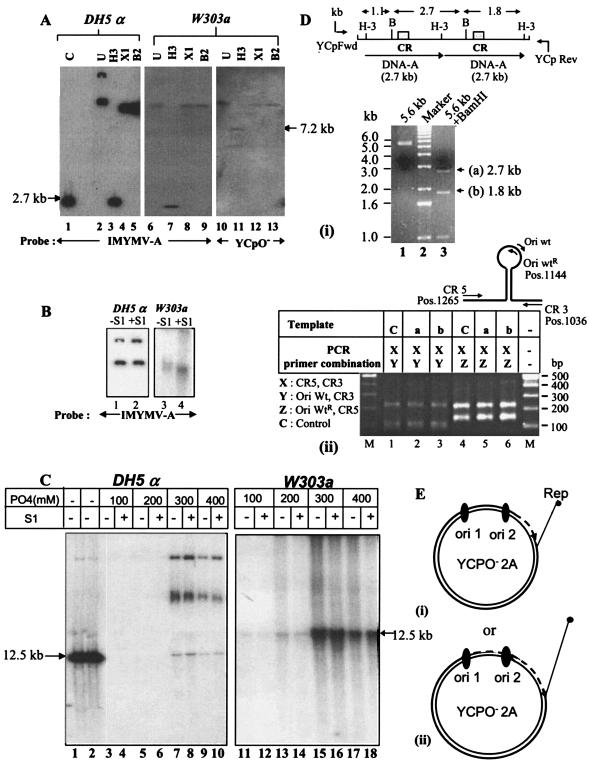
Southern blot and PCR analyses of plasmid products replicated in the W303a yeast strain. (A) Southern blotting of replicated plasmids. Plasmid DNA was isolated from transformed yeast by using a standard protocol (11) or from E. coli DH5α and was treated with a unique cutter, such as HindIII, XhoI, or BglII (H-3, X1, or B2). The digested DNA was resolved by electrophoresis in a 0.7% agarose gel, Southern blotted, and probed as shown. A 2.7-kb IMYMV DNA-A fragment obtained by HindIII digestion of plasmid pUC18-A was used as a length standard (lane 1). (B) Comparison of S1 nuclease sensitivities of plasmid DNAs derived from bacterial and yeast sources. S1 nuclease was used at a concentration of 2 U/μg of DNA at 37°C for 30 min. (C) Characterization of single-strandedness of plasmids replicated in yeast. For further confirmation of the presence of single-stranded DNA, 5 μg of YCpO−-2A plasmid DNA isolated from either E. coli DH5α or S. cerevisiae W303a was allowed to bind separately to mini-HAP columns and step eluted with phosphate buffer (pH 6.8) as indicated. M13 single-stranded DNA and double-stranded RF markers were used as standards during HAP chromatography and were eluted with 75 to 150 mM and 220 to 350 mM phosphates, respectively. The different fractions of eluted plasmids were either mock treated or treated with S1 nuclease. The DNAs of various fractions were sufficiently resolved by agarose gel electrophoresis, Southern blotted, and autoradiographed. The slower migrating bands represent multimeric forms of the 12.5-kb YCpO−-2A. The quantitation of DNA was carried out by measuring the intensities of the 12.5-kb DNA bands in the appropriate lanes with Kodak ID2.0 software. The single-stranded DNA-containing fractions were derived from lanes 11 and 13 (without S1) and 12 and 14 (with S1). Similarly, only the double-stranded DNA fractions from the yeast source were recovered from lanes 15 and 17 (without S1) and 16 and 18 (with S1). The positions of the 12.5-kb DNAs are marked by arrows. (D) Integrity of double copies of DNA-A during replication in yeast. Panel i, the DNA present at the HindIII site of the yeast-derived plasmids was amplified by using primers YCpFwd and YCpRev. The purified 5.6-kb amplified product (lane 1) was digested with BamHI (position 960) to obtain three fragments, of 2.7 kb (a), 1.8 kb (b), and 1.1 kb, respectively (lane 3). The top diagram shows the locations of CRs in the BamHI fragments. Panel ii, fragments a and b containing one origin (CR) were each purified and used as templates for amplification with duplex PCR primer combinations as indicated in the table. For each amplification, two bands of the expected sizes were obtained. The relative positions of the primers employed are shown at the top. The IMYMV DNA-A was used as control template C. (E) Model indicating possible modes of replication of YCpO−-2A plasmid in yeast. We hypothesize that only one of the origins, either ori1 (i) or ori2 (ii), is able to initiate RCR generating full-length YCpO−-2A.