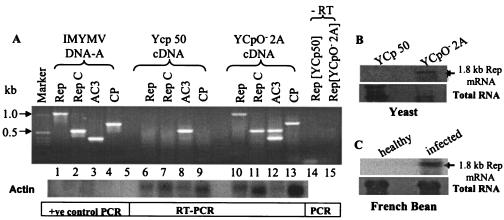
FIG. 3.
Viral transcripts in S. cerevisiae bearing plasmid YCpO−-2A and in infected French bean plants. (A) RT-PCR from RNAs of yeast samples transformed with various plasmids. The PCR products of cDNAs derived from yeast samples (4) bearing YCpO−-2A plasmids are shown for the pairs of primers specific for the Rep (lanes 10 and 11), AC3 (lane 12), and CP (lane 13) genes of IMYMV. RepC is the C-terminal part of Rep. The expected pattern of amplification is shown in lanes 2 to 5 of the positive (+ve) control PCR panel that was obtained from direct amplification from a template of viral DNA-A. The cDNA prepared from the total RNA extracted from YCp50 plasmid-bearing yeast was used as a negative control (lanes 6 to 9). A negative (−RT) control is shown in lanes 14 and 15 indicating that the PCR products are not the artifacts of DNA contamination in the processed RNA samples. A nonspecific band at around 550 bp was observed for both YCp50 control and YCpO−-2A samples in the case of AC3 primers (lanes 8 and 12). Southern blotting with an actin probe was carried out to show uniformity in the loading of cDNA templates (bottom panel). (B) Northern blot analysis of total RNA extracted from the yeast strain W303a bearing the YCpO−-2A plasmid. The presence of a 1.8-kb Rep transcript was revealed by probing with radiolabeled AC1 DNA. Such a transcript was absent from the RNA samples of yeast transformed with the YCp50 control plasmid, as expected. This result is consistent with the observation made with IMYMV-infected French bean plants (C). The total RNAs isolated from the appropriate sources are shown at the bottom for panels B and C.