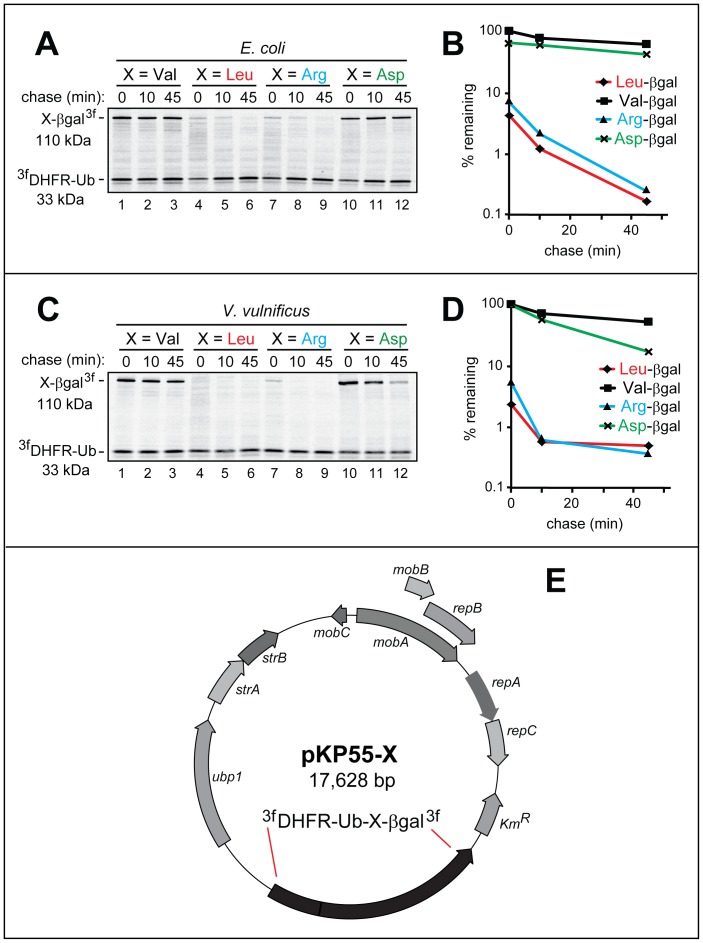
Figure 2. URT pulse-chase assays with model N-end rule substrates in E. coli and V. vulnificus.
The set of URT-based 3fDHFR-UbR48-X-βgal3f fusions (X = Val, Leu, Arg, Asp) was assayed for the in vivo degradation of the released (by the yeast Ubp1 DUB) X-βgal proteins in E. coli (A, B) and in V. vulnificus (C, D) using 35S-pulse-chases (A, C) and their quantification (B, D), as described in Materials and Methods. The bands of the 110 kDa X-βgal test proteins and the 33 kDa 3fDHFR-UbR48 reference protein are indicated on the left. Designations in B and D: squares, Val-βgal; rhombs, Leu-βgal; triangles, Arg-βgal; crosses, Asp-βgal. E. pKP55-X, encoding the S. cerevisiae Ubp1 DUB and 3fDHFR-UbR48-X-βgal3f URT-based fusions. Other notations on the map denote specific bacterial genes. The nucleotide sequences of pKP77 and pKP55-X are available in GenBank (JX181779 and JX181780). In addition, Table S3 contains the nucleotide sequence of pKP55-X.