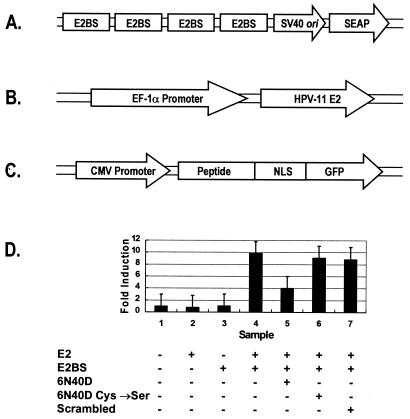
FIG. 2.
Inhibition of E2-mediated reporter gene transcription by 6N40D peptide expressed in Vero cells. (A) Four tandem repeats of the E2-binding-site palindromes (nucleotides 28 to 66 within the HPV-11 origin of replication) were inserted upstream of the minimal simian virus 40 (SV40) promoter and sequence encoding a secreted form of human placental alkaline phosphatase to create the reporter plasmid pSEAP-2. (B) An HPV-11 E2 expression plasmid, pFastBac-E2, was generated by inserting the open reading frame sequence for full-length HPV-11 E2 into pFastBac-EF1αP. Expression from this vector is directed from the EF1α promoter. (C) A DNA cassette encoding the 6N40D E2 inhibitor peptide and a nuclear localization signal peptide (NLS) was inserted into the enhanced GFP fusion vector, pEGFP-N1 (Clontech), and then subcloned into the pFastBac-MAM-1 expression vector, yielding the baculovirus expression construct pFastBac-MAM/6N40D. pFastBac-MAM/6N40D derivative vectors were made which contained the NLS-GFP fusion with double Cys→Ser mutated 6N40D peptide sequence (pFastBac-MAM/6N40DCys→Ser) or the NLS-GFP fusion with a scrambled 6N40D peptide sequence (NH2-SCGDEHMGLECGWVGF-CONH2) (pFastBac-MAM/scrambled). CMV, cytomegalovirus. (D) To test the effects of 6N40D peptides on E2-mediated transcription control in vivo, Vero cells were transiently transfected with 0.125 μg of the reporter plasmid pSEAP-2 and/or 0.5 μg of the E2 transcription activator plasmid pFastBac-E2. The transfected cells were inoculated 24 h later with baculoviruses that expressed 6N40D, double Cys→Ser mutated 6N40D, or scrambled 6N40D-NLS-GFP fusion proteins, according to the bottom panel. Medium supernatant (50 μl) was assayed for SEAP activity 48 h after the transductions. Fold induction represents SEAP activity in Vero cells with mock-transfected cells as reference. At least three independent experiments were performed in triplicate, and the average values are shown. The error bars indicate the standard deviations.