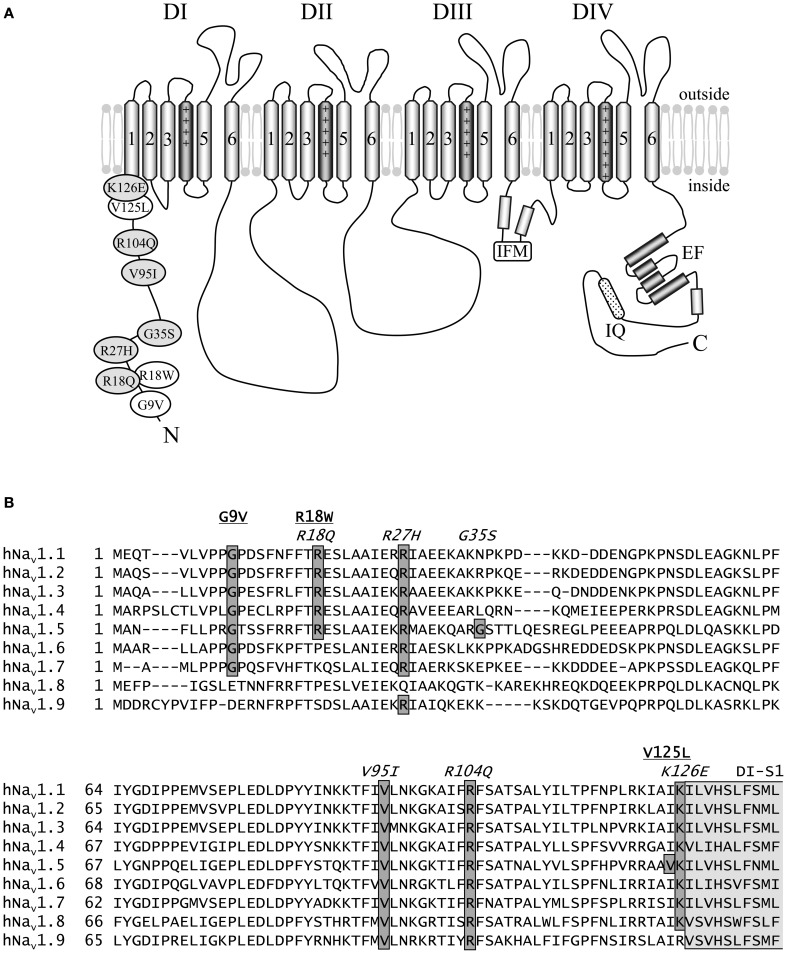
Figure 1.
Schematic representation of hNav1.5 and the N-terminal mutations investigated in this study. (A) Proposed hNav1.5 topology. Affected residues are indicated in white (LQT3) and light grey (BrS). The schematic structure also highlights some important structural features (DI to DIV—domain I to IV, IFM—residues isoleucine, phenylalanine, and methionine of the inactivation gate, IQ—calmodulin binding motif, EF—Ca++ binding EF hand domain). (B) Alignment of the N-terminal sequences of human Nav1.1—Nav1.9. Mutations associated with LQT3 or BrS are underlined or indicated in italics, respectively. Most of the eight affected residues are conserved among the Nav1 subfamily. Residues at position 35 are variable, and position 125 is occupied by isoleucine in all other human Na+ channels. All eight residues affected by a SCN5A mutation are identical in Nav1.5 of human, rat, mouse, and dog (not shown). The N-terminal region of the first putative membrane spanning segment DI-S1 is indicated in light grey. References: G9V (Millat et al., 2006), R18W (Tester et al., 2005), R18Q (Kapplinger et al., 2010), R27H (Priori et al., 2002), G35S (Levy-Nissenbaum et al., 2001), V95I (Liang et al., 2006), R104Q (Levy-Nissenbaum et al., 2001), V125L (Tester et al., 2005), K126E (Vatta et al., 2002a).