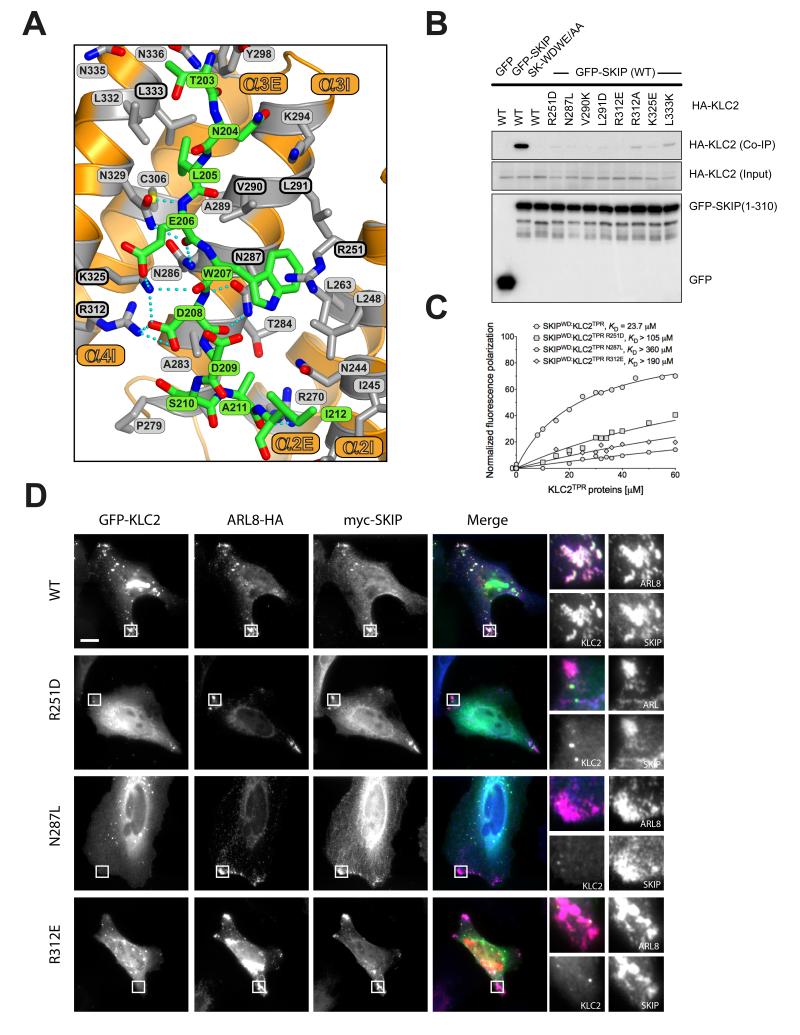
Figure 3.
The SKIPWD-KLC2TPR interface and the effect of KLC2TPR mutations in cargo binding and cellular recruitment. (A) Details of the SKIPWD:KLC2TPR interface. KLCTRP side-chains stabilizing the SKIPWD cargo peptide (green) are shown as grey sticks emanating from the orange cartoon. Non-carbon elements are: nitrogen, blue; oxygen, red; sulfur, yellow. Hydrogen bonds are represented by dotted cyan lines. (B) Co-immunoprecipitation assay showing the effect of KLC2TPR mutations at the SKIPWD:KLC2TPR interface on the interaction. (C) Fluorescence polarization measurements showing that R251D, N287L and R312E mutations in KLC2 dramatically reduce the affinity of the TPR domain for the SKIPWD peptide. KD values reported here were calculated at 150 mM NaCl. (D) Replacement of key KLC2TPR residues results in loss of GFP-KLC2 association with Arl8/SKIP positive lysosomal membranes. Scale bar shows 10μm. In merge panels, GFP-KLC2, Arl8 and SKIP are shown in green, red and blue respectively.