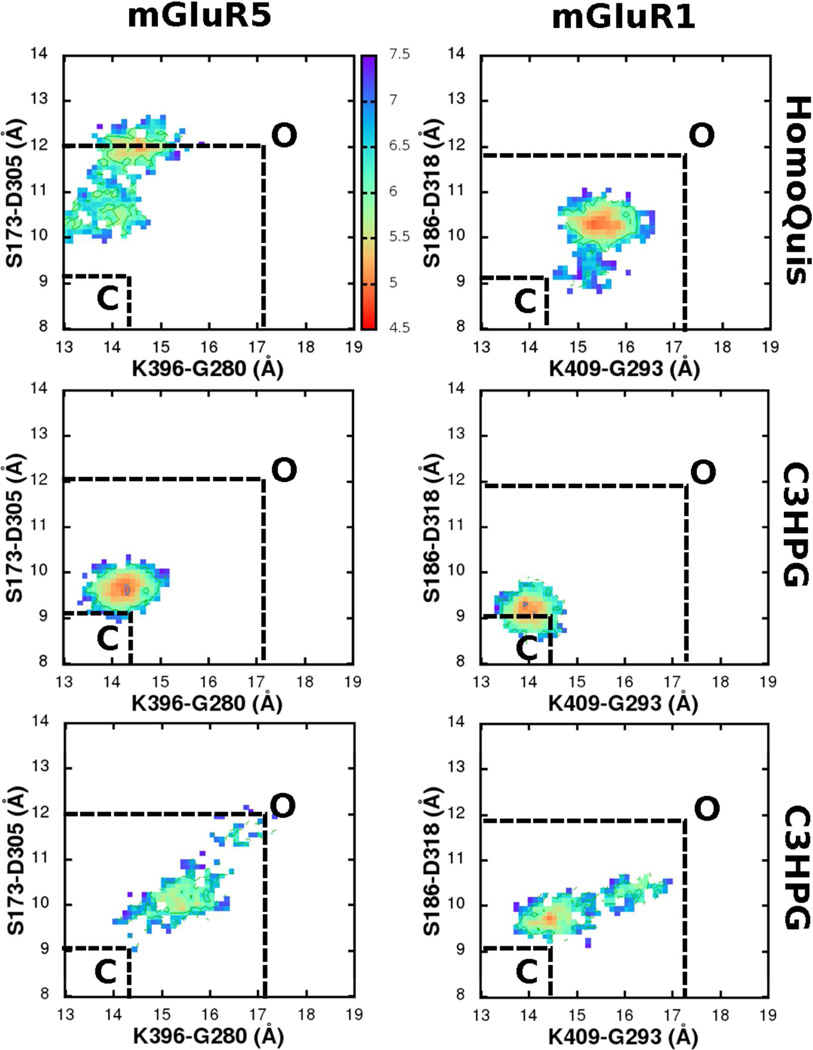
Figure 7.
Comparison of the conformational free energy surfaces of mGluR1 and mGluR5 with HomQuis (HOMQ) and C3HPG ligands in the LBRs. Distances across two pairs of inter-lobe Cα atoms(S173-D305 and K396-G280) are reaction coordinates for the free energies that are measured in kBT; 1 kBT∼0.59 kcal/mol. Closed and open states (C and O) conformational minima are marked based on the free energy profiles from the mGluRx-glutamate and the empty mGluRx (x is 1 or 5) simulations in Figure 5 & Figure 6.