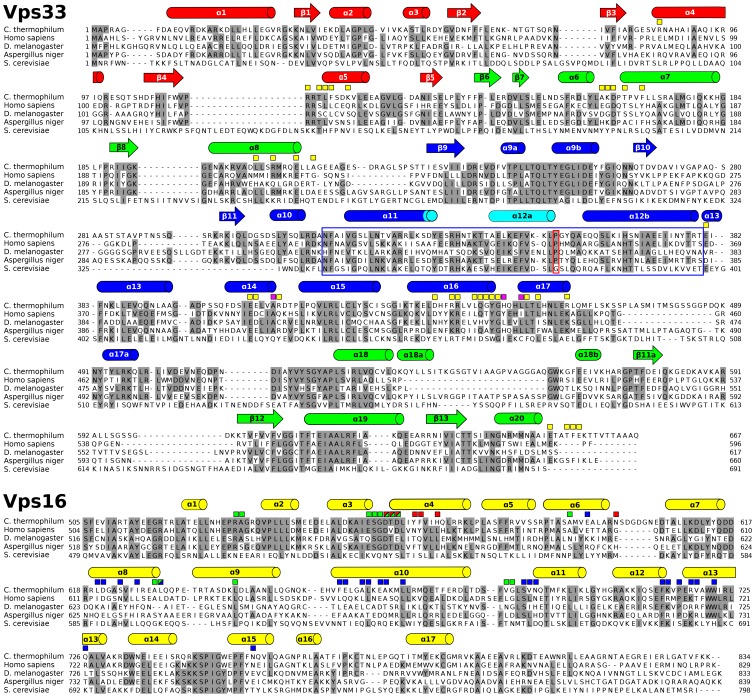
Figure 2. Sequence alignment for Vps33 and the C-terminal region of Vps16.
Intermolecular contacts (<4 Å) are indicated using boxes. For Vps33, these boxes are yellow except for those residues depicted in magenta in Fig. 3B. For Vps16, boxes are color coded to match the Vps33 domain(s) contacted by a given residue. The distal tip of Vps33 domain 3a is highlighted with a blue box. The ‘hinge’ proline (see Fig. 6 legend) is highlighted with a red box. Secondary structural assignments for Vps33 are based on [16] and are colored by domain as in Fig. 1; helix α12 is shown in light blue to indicate that it is ordered in Vps33 but not in Vps16CTD–Vps33 (see text for details). Sequence alignments were performed using CLUSTALW [68] on 15 Vps33 and 15 Vps16 orthologs; for clarity, only 5 orthologs are shown here. The orthologs shown (with percentage sequence identity for Vps33/Vps16 listed in parentheses) are: Homo sapiens (37/33), Drosophila melanogaster (30/27), Aspergillus niger (61/58), and Saccharomyces cerevisiae (19/20).