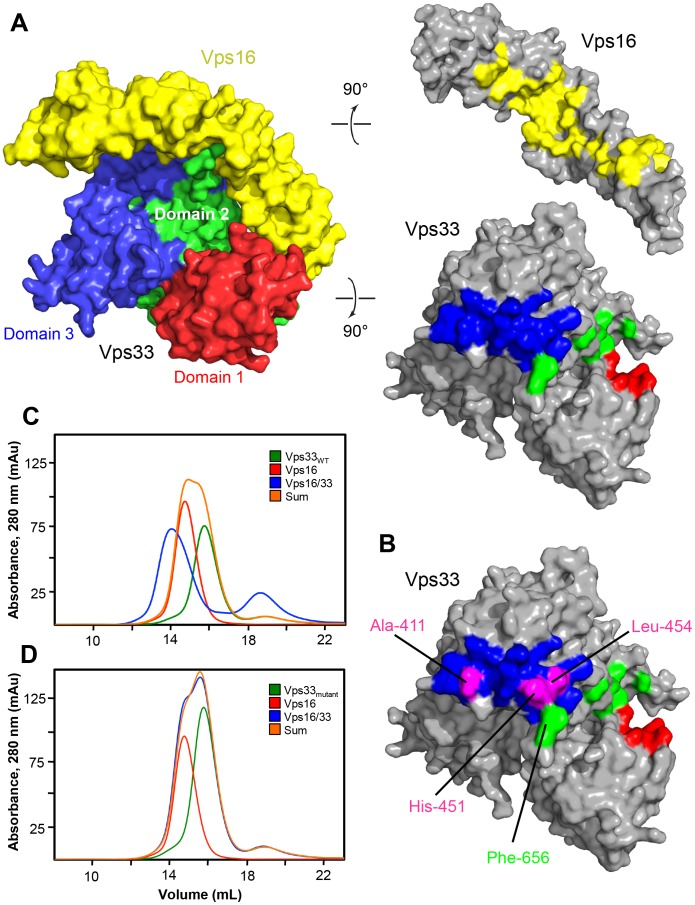
Figure 3. Interaction between Vps16CTD and Vps33.
(A) Vps16CTD and Vps33, oriented as in Fig. 1, are separated and rotated to reveal the contact surfaces. (B) In magenta are shown the positions of Vps33 residue substitutions engineered to disrupt the complex. Also indicated is Phe-656, one of three residues near the C-terminus of Vps33 (and therefore located in domain 2) that is well-ordered only in the Vps16CTD–Vps33 complex. (C) Size exclusion chromatography was used to analyze wild-type Vps33, full-length Vps16, and the combination of the two. Shown for comparison is the sum of the chromatograms for the individual proteins. The Vps16–Vps33 complex elutes earlier from the column, consistent with its larger size. (D) As in panel C, but with Vps33 A411D/H451D in place of wild-type Vps33. The binding reaction is indistinguishable from the sum of the individual protein chromatograms, indicating the absence of a detectable interaction. The same result was obtained for Vps33 A411D/L454E (not shown).