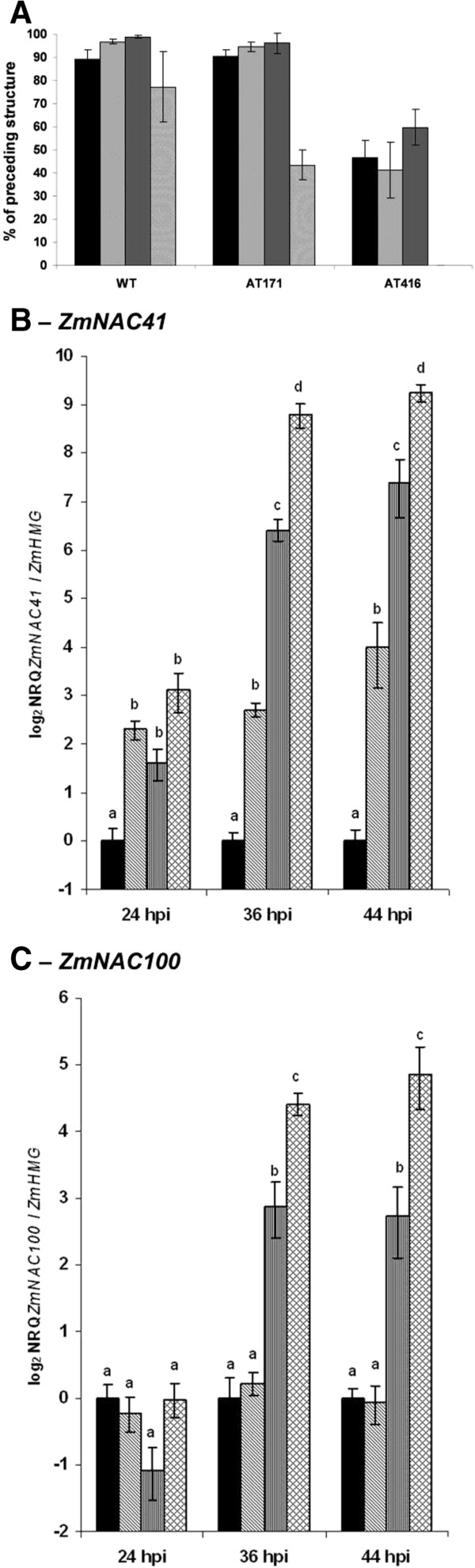
Figure 2.
Induction ofZmNAC41andZmNAC100upon infection withC. graminicolapathogenicity mutants. (A) Germination rates (black bars), appressoria formation (hatched bars), melanisation rates (vertically hatched bars) and penetration rates (crosshatched bars) of C. graminicola CgM2 wild type (left bracket) and the ATMT mutant strains AT171 (middle bracket) and AT416 (right bracket) were assessed at 72 hpi. Four replicate samples per genotype with approx. 100 conidia were assessed and are given ± SE. The data for every developmental stage is given in percent relative to the total number of infection events that exhibited the preceding developmental stage. (B) and (C) Relative quantities of ZmNAC41 (B) and ZmNAC100 (C) transcripts were analyzed by qRT-PCR and are expressed relative to ZmHMG on a log2 scale as means ± SE (n = 4). Mock treated leaves - black bars, wild type strain CgM2 - cross-hatched bars, AT416 mutant - diagonal hatched bars, AT171 mutant - vertical hatched bars. Dissimilar letters indicate significant differences (P-value < 0.05) between the treatments.