Table 3. The gene signature from the NGS dataset using DESeq analysis and their positions in the microarray DEGs by SamR.
| ProbeSetID | GeneSymbol | EnsemblID | log2Ratio | adjustedPvalue | DESeqPosition | SamRposition | SamRlog2FC | SamRq-value(%) |
| 212789 at | NCAPD3 | ENSG00000151503 | 5.58 | 0 | 1 | 96 | 1.23 | 0.00 |
| 204560 at | FKBP5 | ENSG00000096060 | 5.08 | 0 | 2 | 30 | 2.04 | 0.00 |
| 200862 at | DHCR24 | ENSG00000116133 | 3.38 | 0 | 3 | 91 | 1.14 | 0.00 |
| 205876 at | LIFR | ENSG00000113594 | 4.24 | 4.72E-307 | 5 | 374 | 0.68 | 0.89 |
| 219555 s at | CENPN | ENSG00000166451 | 5.09 | 2.12E-295 | 6 | 29 | 1.93 | 0.00 |
| 222118 at | CENPN | ENSG00000166451 | 5.09 | 2.12E-295 | 6 | 11 | 2.58 | 0.00 |
| 218211 s at | MLPH | ENSG00000115648 | 2.69 | 1.29E-282 | 7 | 913 | 0.47 | 3.09 |
| 203455 s at | SAT1 | ENSG00000130066 | 2.93 | 1.15E-193 | 10 | 146 | 0.95 | 0.32 |
| 210592 s at | SAT1 | ENSG00000130066 | 2.93 | 1.15E-193 | 10 | 161 | 0.91 | 0.32 |
| 213988 s at | SAT1 | ENSG00000130066 | 2.93 | 1.15E-193 | 10 | 148 | 0.95 | 0.32 |
The list of identifiers and their associated genes extracted from the NGS dataset using DESeq analysis and put to the sscMap. We established where these genes were located in full list (Table S3) of statistically differentially expressed genes returned by the SamR analysis on the microarray dataset. All these genes lay within a SamR reported FDR of 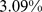 . Table S4 also contains the signed ranks of these 10 probesetIDs in the 6 instances of reference profiles for cotinine.
. Table S4 also contains the signed ranks of these 10 probesetIDs in the 6 instances of reference profiles for cotinine.
