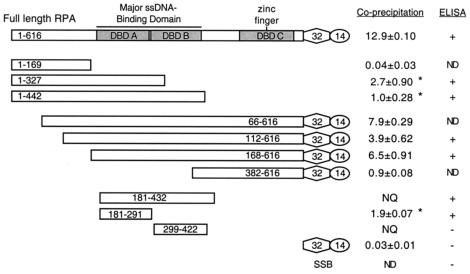
FIG. 1.
Summary of the RPA-E1 interaction mapping studies. Full-length RPA and RPA70 truncation mutants were tested for their ability to interact with BPV1 E1 in GST affinity coprecipitation and ELISA-based protein interaction assays. In this figure, RPA70 is shown as long rectangles, RPA32 is shown as hexagons, and RPA14 is shown as circles. The ssDNA-binding domains within RPA70 are shaded and indicated as DBD-A, DBD-B, and DBD-C. Together, DBD-A and DBD-B form the primary ssDNA-binding region within RPA. A fourth ssDNA-binding domain, DBD-D, is located within RPA32. The zinc finger domain, which is embedded in DBD-C, is also indicated. Since the carboxyl terminus of RPA70 is essential for heterotrimer assembly, mutants containing the carboxyl terminus were expressed in a complex with RPA32 and RPA14. The results for the coprecipitation assays were taken from at least three separate experiments. The numbers indicate the percentages of 35S-RPA precipitated with GST-E1 beads (± standard deviation), after the subtraction of background (binding of that protein to GST Sepharose beads). Binding to E1 that could not be assessed due to excessive proteolysis of the 35S-labeled RPA protein is labeled as NQ (not quantifiable). (*, two N-terminal RPA domain constructs, RPA70(1-327) and RPA70(1-442), clearly showed much higher levels of coprecipitation with GST-E1 than did the RPA70(1-169) construct, but the levels of coprecipitation were lower than those for the C-terminal constructs. This was attributable to a relatively higher degree of proteolysis for the N-terminal domain constructs than for the C-terminal domain constructs.) Results from the ELISA are summarized as binding comparable to that of full-length RPA (+) and binding comparable to that of the negative control, E coli SSB (−). Mutant proteins that were not tested in a particular assay are noted as ND (not done).