FIG. 2.
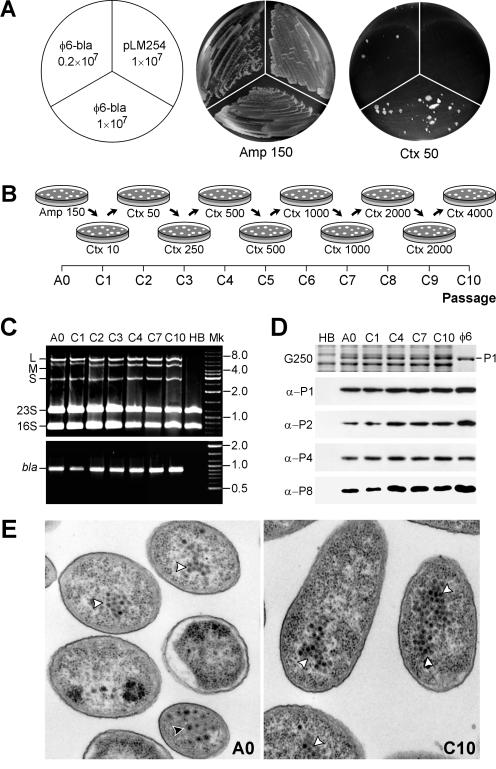
φ6 bla carrier cells rapidly adapt to CTX. (A) HB10Y(φ6-bla) carrier-state cells (0.2 × 107 to 1 × 107) were plated onto LB agar containing either 150-μg/ml AMP (Amp150) or 50-μg/ml CTX (Ctx50). CTX-resistant colonies appeared after 3 days of incubation at 28°C. No colonies were detected at this time on the sector inoculated with 107 HB10Y(pLM254) cells, which contain a plasmid encoding the bla gene. (B) Schematic diagram of the CTX adaptation experiment. Cells were cultivated on LB agar containing increasing CTX concentrations (in micrograms per milliliter), as shown below the diagrams. Twenty to 40 of the largest colonies were pooled after each passage and were used for subsequent rounds of selection. (C) Agarose gel analysis of RNAs extracted from carrier-state cells at passages A0, C1, C2, C3, C4, C7, and C10 (top panel). HB, RNA from uninfected HB10Y cells. Also shown are RT-PCR products generated with bla-specific primers (bottom panel). Other designations are defined as for Fig. 1. (D) SDS-polyacrylamide gel electrophoresis analysis (29) of carrier-state cells from different passages (A0, C1, C4, C7, and C10) or of purified φ6 virus (φ6). HB, uninfected HB10Y cells. Panels: G250, a Coomassie G250-stained gel fragment showing the band of protein P1; α-P1, α-P2, α-P4, and α-P8, immunoblots produced with antibodies specific to corresponding φ6 nucleocapsid proteins and visualized by ECL detection (Pierce Biotechnology). (E) Transmission electron micrograph of osmium tetroxide- and uranyl acetate-stained thin cell sections from A0 and C10 passages produced as described previously (2). Black arrowhead, enveloped virions; white arrowheads, nucleocapsids or PC particles.