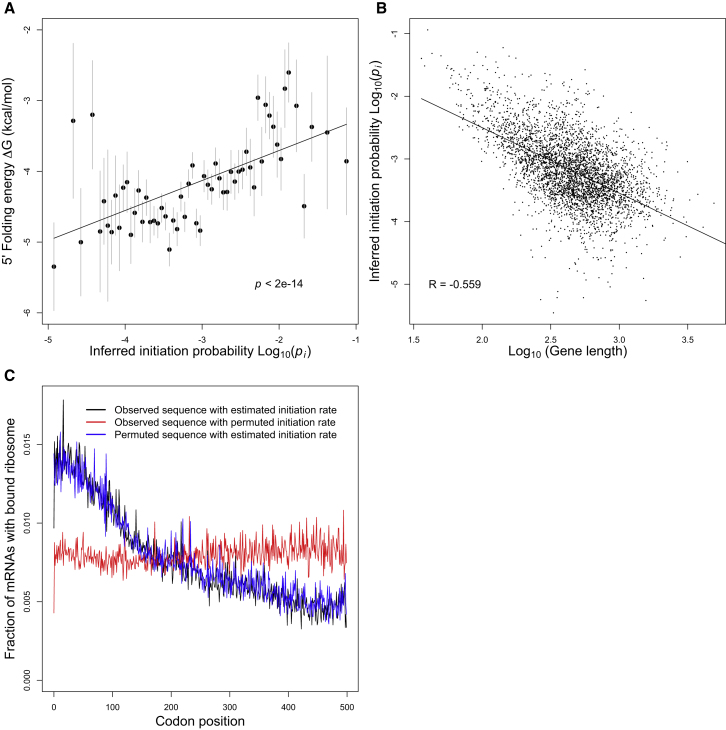
Figure 5.
Fast Initiation of Short Genes Causes a 5′ Ribosomal Ramp
(A) Yeast genes with weak 5′ mRNA structure initiate quickly. We inferred the initiation probabilities of 3,795 endogenous yeast genes from ribosomal profiling data (Ingolia et al., 2009). A gene’s initiation probability correlates strongly with the estimated energy of its 5′ mRNA structure. The gray bars indicate 1 SD of folding energies of binned genes.
(B) Initiation probabilities of yeast genes also correlate with ORF lengths, suggesting that short genes have experienced selection for faster initiation.
(C) Simulations of translation in a wild-type yeast cell recapitulate the “ramp” of 5′ ribosomes observed in empirical ribosomal profiling data (Ingolia et al., 2009). The figure shows the density of ribosomes bound to mRNAs as a function of codon position, averaged across the simulated transcriptome (black). The ramp is preserved in simulations that permute the codons within each gene (blue), but the ramp is disrupted when permuting the initiation probabilities among genes (red). Thus, we infer that the ramp of 5′ ribosomes is caused primarily by the trend toward faster initiation in short genes, rather than by the ordering of codons within each gene.
See also Figures S3, S4, and S5.