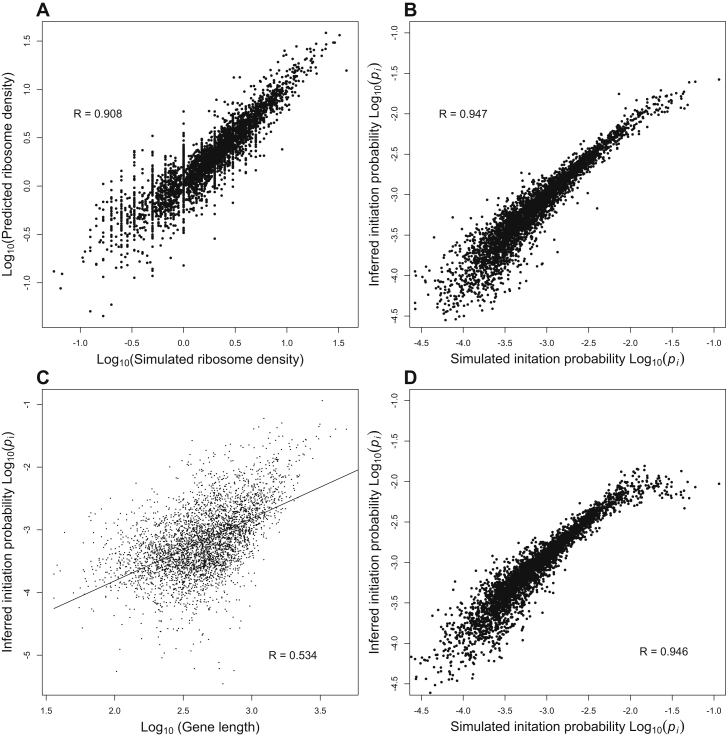
Figure S1.
Validation of Analytical Approximations of Simulations Model, Related to Figure 1
(A) We have derived analytic approximations for the steady-state density of ribosomes on each mRNA molecule. These equations provide a good approximation to the true behavior of our stochastic simulation.
(B) We can invert these equations to infer the initiation probability of each gene, based on its observed equilibrium ribosome density. Panel B shows a validation experiment in which we applied this inference method to data that had been simulated with known initiation probabilities, confirming that we can accurately infer these parameters.
(C) Correlation between gene length and initiation probabilities does not bias our inference of initiation probabilities. We validated that we can reliably infer initiation probabilities from observed ribosome densities, even when gene length and initiation probabilities are positively correlated. To do so we inverted the rank order of initiation probabilities assigned to yeast genes, so that the genes with longer ORFs were now assigned greater initiation probabilities.
(D) When we simulated a cell with this new assignment of initiation probabilities we were still able to reliably infer the simulated initiation probabilities based on the simulated ribosome density for each gene. These results confirm that the negative correlation between gene length and inferred initiation probability observed in the real data (main text Figure 5B) is not an artifact of our inference procedure.