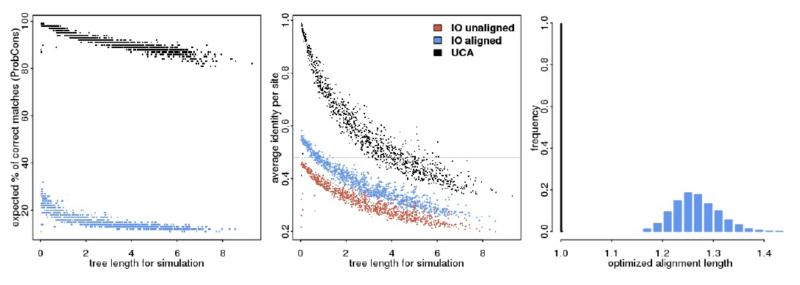
Figure 3.
Alignment properties for data sets simulated under Universal Common Ancestry (UCA) and Independent Origins (IO). The left panel shows the relation between quality values from ProbCons (averaged over all sites) and tree length used in simulation. The middle panel shows the average pairwise identity under UCA and IO as a function of tree length. For IO simulations we have the identity values before and after optimizing the alignment, and the gray horizontal line at 0.47 represents the observed value for the BE data set. The panel at the right show the histogram of optimal alignment lengths divided by the unaligned values, for simulations under UCA and IO. Here the distribution is pooled over several tree lengths. For all panels we have the UCA simulations in black (after alignment optimization, which is essentially the same as before optimization), the unaligned IO data sets in red and after alignment optimization in blue.