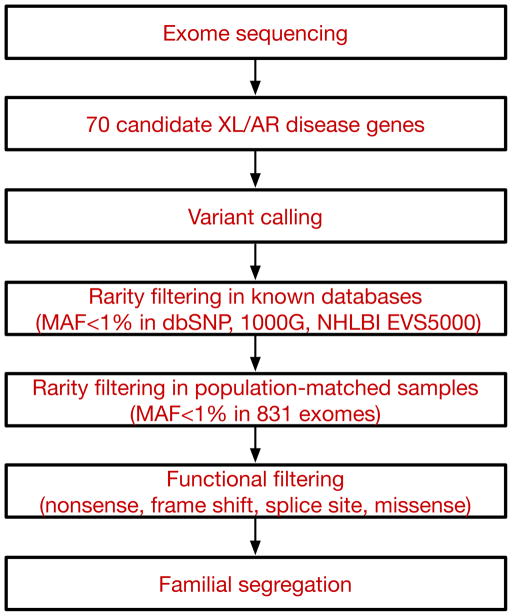
Figure 4.
Schematic of the analytical pipeline. Tiered strategy for examining the cohort of ASD families for inherited mutations in known disease genes. Whole exome sequencing data was used to systematically survey 70 genes (listed in Table S2) known to be associated with autosomal recessive or X-linked neurodevelopmental illness and autistic features. Variants were filtered based on rarity (MAF<1%) in known databases and our internal, population-matched dataset (see Experimental Procedures for details), and predicted functional effect on the protein. All candidate variants were validated by Sanger sequencing and were required to segregate within the family. XL: X-linked, AR: autosomal recessive, MAF: minor allele frequency, 1000G: 1000 Genomes Project, NHLBI EVS5000: NHLBI Exome Sequencing Project.