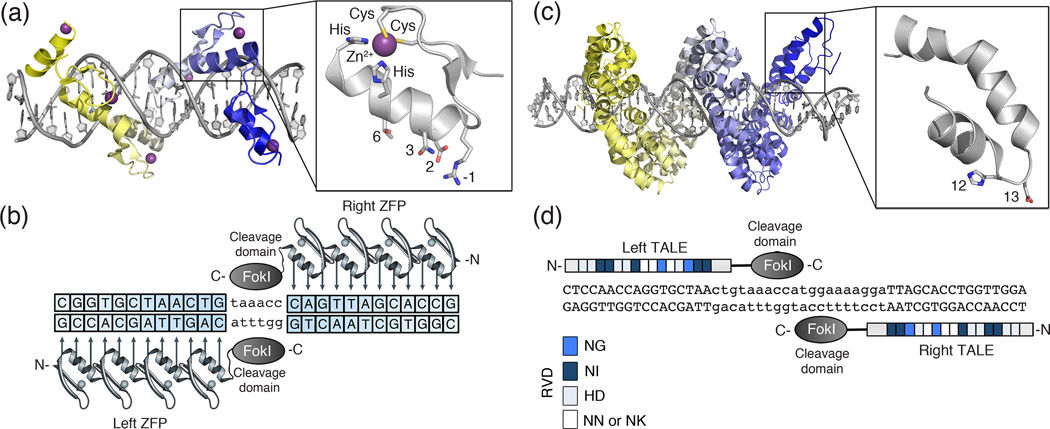
Figure 1. Structure of zinc-finger and transcription activator-like effectors.
(a) (Top) Designed zinc-finger protein in complex with target DNA (grey) (PDB ID: 2I13). Each zinc-finger consists of approximately 30 amino acids in an ββα arrangement (inset). Surface residues (−1, 2, 3 and 6) that contact DNA are shown as sticks. Each zinc-finger domain contacts 3–4 base pairs (bps) in the major groove of DNA. The side chains of the conserved Cys and His residues are depicted as sticks in complex with a Zn2+ ion (purple). (b) Cartoon of a zinc-finger nuclease (ZFN) dimer bound to DNA. ZFN target sites consist of two zinc-finger binding sites separated by a 5- to 7-bp spacer sequence recognized by the FokI cleavage domain. Zinc-finger proteins can be designed to recognize unique “left” and “right” half-sites. (c) (Top) TALE protein in complex with target DNA (grey) (PDB ID: 3UGM). Individual TALE repeats contain 33–35 amino acids that recognize a single bp via two hypervariable residues (repeat-variable diresidues; RVDs) (shown as sticks) (inset). (d) Cartoon of a TALE nuclease (TALEN) dimer bound to DNA. TALEN target sites consist of two TALE binding sites separated by a spacer sequence of varying length (12-to 20-bp). TALEs can be designed to recognize unique “left” and “right” half-sites. RVD compositions are indicated.