Fig. 1.
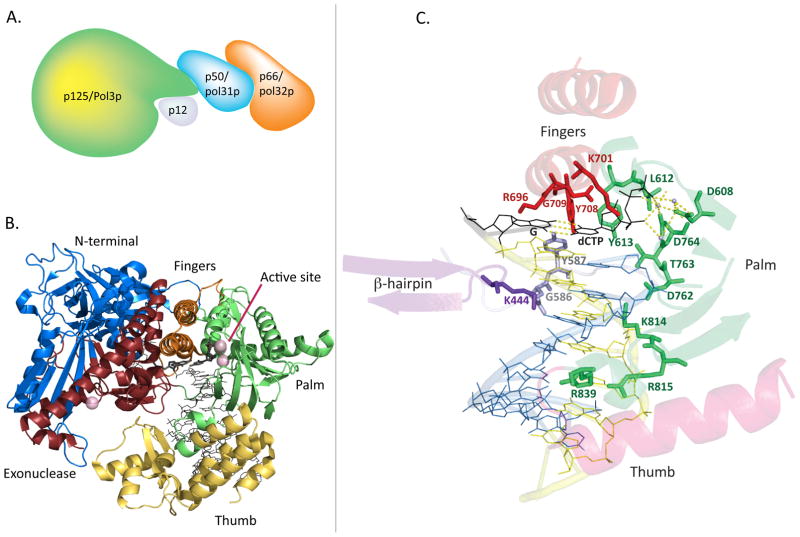
Structure of the Pol δ holoenzyme, catalytic subunit, and DNA binding pocket. A. A conceptual depiction of the four-subunit human Pol δ holoenzyme, based on demonstrated interactions of subunits and a small-angle X-ray scattering study (see text). B. Cartoon representation of the crystal structure of the p125 catalytic subunit in complex with DNA and the incoming dCTP (black) bound at the active site. Ca2+ ions are shown as purple spheres, representing the location of the Mg2+ atoms at the polymerase and exonuclease active sites. C. Pol δ active site and DNA binding channel, highlighting important side chains for polymerase fidelity, as well as purported “sensing” side chains along the minor groove. Palm residues are green, fingers residues are red, N-terminal domain residues are silver, β-hairpin site is purple, DNA template strand is yellow, and DNA primer strand is blue. The incoming dCTP and its template G are shown in black, and active site metals are shown as light blue spheres. Hydrogen bonds (yellow) are shown for the nascent base pair and for the active site metals. (Structure images generated in The PyMOL Molecular Graphics System, Version 1.5.0.4 Schrödinger, LLC. from PDB accession code 3IAY).